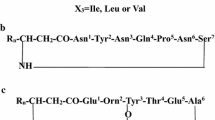Abstract
Mass spectrometric analysis of a banyan endophyte, Bacillus subtilis K1, extract showing broad spectrum antifungal activity revealed a complex mixture of lipopeptides, iturins, surfactins, and fengycins. Fractionation by reversed-phase high performance liquid chromatography (HPLC) facilitated a detailed analysis of fengycin microheterogeneity. Matrix assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) mass spectrometric studies permitted the identification of several new fengycin variants. Four major sites of heterogeneity are identified: (1) N-terminus β-hydroxy fatty acid moiety, where chain length variation and the presence of unsaturation occur, (2) position 6 (Ala/Val/Ile/Leu), (3) position 10 (Val/Ile) within the macrocyclic ring, and (4) Gln to Glu replacement at position 8, resulting in fengycin variants that differ in mass by 1 Da. Diagnostic fragment ions provide a quick method for localizing the sites of variation in the macrocycle or the linear segment. Subsequent establishment of the sequences is achieved by MS/MS analysis of linear fengycin species produced by hydrolysis of the macrocyclic lactone. Unsaturation in the fatty acid chain and the presence of linear precursors in the B. subtilis K1 extract are also established by mass spectrometry. The anomalous distribution of intensities within isotopic multiplets is a diagnostic for Gln/Glu replacements. High resolution mass spectrometry facilitates the identification of fengycin species differing by 1 Da by localizing the variable position (Gln8/Glu8) in the fengycin variants.
Similar content being viewed by others
1 Introduction
Microorganisms possess a remarkable biosynthetic ability to produce metabolites with diverse biological activities. Endophytic organisms, bacteria, and fungi often play a very important role in the defense mechanisms of their host plants against pathogens [1, 2]. Traditional medicines based on plant extracts may indeed exploit the therapeutic properties of metabolites produced by both host plants and their endophytes. The banyan tree (Ficus benghalensis), an important Asian plant, is characterized by an extremely long life span and numerous anecdotal accounts of its use in traditional medicine [3]. The majority of the endophytes isolated from woody trees are fungi [1, 2, 4]. Various forms of ascomycete and mitosporic fungi have been isolated from aerial roots of the banyan tree [5]. To our knowledge, a detailed characterization of metabolites from bacterial endophytes from this source has not been undertaken. We, therefore, investigated the biological activity of seven banyan endophytic Bacilli isolated from aerial roots that exhibited antifungal activity. Of these seven isolates, Bacillus subtilis K1 exhibited potent as well as broad spectrum fungal antagonistic activity. The molecular characterization of the extract of B. subtilis K1 is described in this report.
A great deal of interest has centered around the diversity of the peptides produced by non-ribosomal peptides synthetases (NRPSs) [6–8]. Bacillus subtilis and other related bacillus species have been shown to be extremely prolific in the production of lipopeptides under culture conditions [9–12]. The interest in lipopeptides largely stems from their potential as biocontrol agents for mitigating plant diseases [13–15]. In previous studies, lipopeptides of the surfactin, iturin and fengycins families from various Bacilli species, including B. subtilis, B. amyloliquefaciens, B. licheniformis, B. globigii, B. pumilus, B. cereus, B. megatarium, and B. thurigiensis have been characterized [10–12, 16–32]. Extensive microheterogeneity is observed in all these products of non-ribosomal polypeptide synthesis [11, 20, 22, 23]. The iturins are eight residue cyclic peptides containing an unusual β-amino fatty acid moiety [10, 17, 18, 33, 34]. Mycosubtilins and bacillomycins characterized from B. subtilis strains belong to the iturin family [28, 29]. The surfactins and fengycins are cyclic depsipeptides possessing a hydrolysable ester linkage [6, 19, 20]. The lichenysin from B. licheniformis and pumilacidin from B. pumilus are related to surfactin [31, 32]. Fengycins are cyclic depsipeptides formed by lactonization involving the side chain -OH of Tyr3 and C-terminal -COOH of Ile10. The fengycins posses a three residue tail segment (Scheme 1a) [19]. The plipastatins from B. cereus are related to fengycins, differing in the configurations of the two tyrosines [21, 27]. Two classes of fengycins, A and B, which differ by an Ala to a Val change at position 6, have been reported thus far [19]. Tandem mass spectrometric methods have been extensively used to characterize fengycins [11, 12, 22–24]. The peptide sequences of fengycin variants from various Bacilli strains have been determined using intact as well as partially hydrolyzed fengycins by matrix assisted laser desorption ionization (MALDI) mass spectrometry and liquid chromatography coupled with electrospray ionization tandem mass spectrometry (LC-ESI-MS) [11, 12, 22, 25]. Fengycins A and B have been identified on the basis of characteristic intense fragment ions [22]. Besides these two classes, further heterogeneity in the fengycins is observed in the length of the β-hydroxy fatty acids (C14-C17) [11, 12, 19, 22–25]. The presence of unsaturated β-hydroxy fatty acids in the fengycins has also been reported previously [11, 19, 25]. As a consequence, these lipopeptides are produced as extremely complex mixtures of closely related molecules in which diversity is generated by variations in both the fatty acid and peptide components of the structures [6, 8, 11]. The application of LC-ESI-MSn methods permits deconvolution of these complex peptide libraries, facilitating identification of new lipopeptide variants. The results of the present study establish the sequences of several new fengycin variants.
2 Experimental
2.1 Microorganisms
Seven endophytic Bacilli designated as B. subtilis K1, B. subtilis A2, B. subtilis A4, B. subtilis A12, B. amyloliquefaciens A11, B. amyloliquefaciens A13, and Bacillus sp. A32 were isolated from aerial roots of the banyan tree by surface sterilization followed by cultivation on Luria Bertani agar (LA) medium. All seven cultures were maintained on LA slopes at 4 °C, as well as in the form of glycerol stocks at –20 °C. All seven isolates were tested for in vitro antagonism against 11 pathogens (Aspergillus niger 1142, Alternaria brunsii, Fusarium oxysporum, Aspergillus parasiticus, Aspergillus flavus, Cladosporium herbarum 1112, Lasiodiplodia theobromae ABFK1, Candida albicans, Trichosporon 1110, Mucor indicus, and Chrysosporium indicum). Among these isolates, B. subtilis K1 exhibited the maximum antifungal activity against all the test fungi, and was selected for production as well as separation of lipopeptides. The 16 S rDNA sequence of B. subtilis K1was determined at GeneI, Bangalore. The full length sequence of 16 S rDNA (1515 kb) of B. subtilis K1was submitted to Genebank and the accession number obtained was EU056571 [35].
2.2 Production and Extraction of Lipopeptides
For the production of lipopeptides, cells from a single colony of B. subtilis K1 were inoculated in 50 mL sterile Luria Bertani (LB) broth (Himedia, Mumbai, India) in a 250 mL Erlenmeyer flask, incubated at 30 °C for 12 h (O.D. 1.9–2.0) on an orbital shaker (150 rpm) and used as an inoculum. An appropriate aliquot of inoculum was added to 250 mL of sterile glucose yeast extract (GY) medium (composition g/L: glucose, 10.00; yeast extract, 1.00; KH2PO4 2H2O, 1.00; K2HPO4 2H2O, 1.00; MgSO4. 7H2O, 0.20; CaCl2 2H2O, 0.02; FeSO4.2H2O, 0.05) in 1000 mL Erlenmeyer flasks to obtain an initial O.D. (600 nm) ~0.05. The flasks were incubated on an orbital shaker (150 rpm) at 30 °C for 72 h. The cells were separated by centrifugation (~10,000 × g for 20 min) and lipopeptides from culture supernatant were precipitated by lowering the pH of the broth to 2 using 6 N HCl. The precipitates were harvested by centrifugation of the acidified broth at ~10,000 × g for 20 min. The supernatant was discarded while the pellet was solubilized in pure methanol. The methanolic extract was then centrifuged to remove the undissolved fraction, while the supernatant was collected and dried by a rotary vacuum evaporation at 30 ºC. The crude extract, obtained as a yellowish brown, gummy material was used for further separation and characterization.
2.3 Separation of Lipopeptides by HPLC
The lipopeptides in the crude extract were further separated by reversed phase high performance liquid chromatography (RP-HPLC), using a semi-preparative Phenomenex (Torrance, CA) C18 column (4.6 mm × 250 mm, 10 m particle size, 90 pore size) and MeOH/ H2O/ 0.1 % trifluoroacetic acid (TFA) as a mobile phase. The flow rate was maintained at 1 mL/min with a gradient of 60 min (80 %–95 %, vol/vol MeOH in 50 min; 95 %, vol/vol MeOH for 5 min, and 95 %–80 %, vol/vol MeOH in 5 min). The elution of metabolites was monitored using UV detection at 226 and 280 nm. The metabolites eluting under individual peaks were separately collected in different vials, concentrated, lyophilized, and used for further mass spectrometric characterization.
2.4 Chemical Modifications for Structure Determination
2.4.1 Saponification
The HPLC fractions were mixed in equal proportion with 2 M sodium hydroxide (NaOH) and allowed to react overnight at room temperature. The reaction was terminated by addition of a few drops of 100 mM glacial acetic acid to the reaction mixture so as to lower the pH to 2. The saponified peptides were extracted with an equal volume of ethyl acetate, three times. All ethyl acetate fractions were pooled and solvent was evaporated using a SpeedVac (Remi, Mumbai, India). The residue was solubilized in methanol and excess NaOH was removed by passing the solution through a ZipTip C18 (Millipore, Mumbai, India). The trapped sample was washed twice with 0.1 % TFA, and then eluted from the cartridge with 100 % (v/v) of MeOH in 0.1 % TFA. For mass spectrometry, TFA was removed through evaporation and the sample was dissolved in a required amount of MeOH in 0.1 % (vol/vol) acetic acid.
2.4.2 Esterification
The esterification of lipopeptides was carried out by incubating the 1 μL of HPLC fraction with 50 μL benzyl alcohol-HCl (prepared by the drop wise addition of 200 μL of acetyl chloride to 1 mL of benzyl alcohol) for 30 min at 37 °C. The HCl was removed through evaporation and the esterified sample was further analyzed by mass spectrometry. To monitor the esterification reaction over a period of time, each sample was incubated with benzyl alcohol-HCl (1:50 vol/vol) for 0 to 10 h and analyzed by mass spectrometry.
2.5 Mass Spectrometry
An Ultraflex TOF/TOF MALDI mass spectrometer (Bruker Daltonics, Billerica, MA and Bremen, Germany) was used to acquire MALDI mass spectra in positive ion mode, within a mass range 550–2000 m/z. For MALDI-MS analysis, HPLC fractions and crude methanolic extract from B. subtilis K1 were mixed with α-cyano-4-hydroxy-cinnamic acid saturated in acetonitrile: water (1:1 vol/vol) containing 0.1 % (vol/vol) trifluoroacetic acid in equal amount and applied on the MALDI sample plate and left to air dry. For MALDI-MS/MS analysis, the spectra were recorded using a LIFT option in Ultraflex II TOF/TOF by selecting appropriate precursor ions within a window range of ±3 Da. The Flexanalysis software (Bruker Daltonics, ver. 3.2) was used for the MALDI-MS data analysis.
For the intact cell MALDI-MS analysis, cells from a single colony of B. subtilis K1 grown on sterile LA plates (at 30 °C for 72 h) were transferred into 20 μL of methanol: water (1:1) and 1 μL of bacterial cell suspension was used for MALDI-MS analysis.
The LC-ESI MS experiments were performed on a micrOTOF Q-II (Bruker Daltonics, Bremen, Germany) mass spectrometer coupled with an Agilent 1100 HPLC (Agilent Technologies, CA, USA). Water-acetonitrile (with 0.1 % formic acid) was used as the solvent system. The LC gradient was set from 80 % acetonitrile to 100 % acetonitrile over 50 min, in a linear gradient, at a flow rate of 0.2 mL/min. The separation was performed on a C18 RP analytical column (4.6 × 150 mm, 2.7 μm particle size). The mass spectra were acquired at 1 Hz scan rate, with three most abundant ions of each scan being subjected to MS/MS analysis. For auto MS/MS, collision rf was ramped from 800–400 Vp-p and the collision energy (CE) was ramped from 80 % to 120 % of the base values of the collision energy for the precursor. In the acquisition software, the base value of the collision energy for the singly charged m/z of 1000, 1500, and 2000 were set at 35, 50, and 70 eV, respectively. The CE base values for the precursor, with m/z value within this range, were auto-calculated from these values linearly. The source temperature and voltage were set at 525 K and 4500 V, respectively. The dry gas flow and nebulizer pressure were set at 8.0 L/min and 27 psi, respectively. LC-ESI-MS experiments were also carried out on a HCT Ultra ETD II (Bruker Daltonics, Bremen, Germany) ion trap mass spectrometer coupled with an Agilent 1100 HPLC system, under elution conditions described above. For HPLC fractions, MS and MS2 spectra were acquired by direct infusion using a syringe pump (Cole-Parmer, Vernon Hills, IL, USA) pumping at 200 μL/h flow rate. The ESI-MS data were processed using Data Analysis software ver. 4.0 (Bruker Daltonics, Bremen, Germany).
3 Results and Discussion
Figure 1a shows the MALDI mass spectrum obtained from a cell suspension of Bacillus subtilis K1. Two groups of molecular ions in the m/z range 1000–1200 and 1400–1550 are observed. The former represents a mixture of surfactins and iturins, whereas the latter may be assigned as fengycins. Figure 1b shows the UV detected HPLC trace of the B. subtilis K1 extract. Individual fractions were mass spectrometrically analyzed in order to establish the presence of lipopeptide components, which were anticipated to have masses greater than 900 Da. Peak P1, which has the shortest retention time, contains substances with intense UV absorbance at 280 nm and lacks peptides with masses greater than 900 Da. All the remaining peaks (P2-P23) yielded mass spectra establishing the presence of peptidic components in the mass range, expected from iturins (m/z 1028–1073), fengycins (m/z 1421–1566), and surfactins (m/z 994–1065) [11, 18–24]. The fengycin family could be readily distinguished by its distinctly higher masses in the range of m/z 1421–1566 [11, 19, 22–24]. MS/MS analysis of each molecular ion from HPLC peaks P2-P8 and P20-P23 established the identification of iturin A1 to iturin A7 and C13-C17 surfactin variants, respectively. All the identified iturins and surfactins correspond to lipopeptides previously reported in the literature [11, 17, 18, 20, 23, 33, 34]. The present study, therefore, focuses on mass spectral characterization and identification of new fengycins.
(a) MALDI-MS spectrum a B. subtilis K1 cell suspension, S-surfactin, I-iturin, F-fengycin and (b) elution profile of lipopeptides separated from a crude methanolic extract (obtained from cell free fermentation broth of B. subtilis K1) using a semi-preparative reversed phase C18 HPLC column (4.6 mm × 250 mm, 10 m particle size, 90 pore size). The m/z values of the most abundant molecules in HPLC fractions (P1-P23) are shown above the peaks. (Inset) MALDI-MS spectrum of a methanolic crude extract of B. subtilis K1. The spectrum shows a metabolite distribution similar to that obtained from intact cells (a)
3.1 Mass Spectrometry of Fengycins and Identification of Three New Classes
Peaks P6 and P8-P19 (Figure 1b), yielded masses in the range (m/z 1421.8–1566.8) anticipated for fengycins and were subjected to subsequent MS/MS analysis.
3.2 Fengycin A (Ala6, Ile10) and Fengycin B (Val6, Ile10)
Scheme 1a shows a fengycin structure which consists of a cyclic peptide lactone formed between the phenolic -OH of Tyr3 and the C-terminal -COOH of an amino acid at position 10. The N-terminus of the decapeptide segment is acylated with a β-hydroxy fatty acid (β-OH FA). Two classes of fengycins have been reported thus far. Fengycin A has DAla at position 6, while fengycin B has DVal (Scheme 1a). Within each class, further heterogeneity is observed corresponding to the presence of a β-OH FA, with the chain length varying from C14–C19 [11, 21–24]. Under collision induced dissociation (CID) conditions, intense product ions corresponding to cleavage of Glu1-Orn2 and Orn2-Tyr3 are readily observed and have been used as diagnostics for identifying fengycin variants with the amino acids replacement within the macrocyclic moiety, (Scheme 1a). Figure 2a–d show ESI-MS/MS spectra obtained from HPLC fractions-P8, P15, P16, and P17, which appeared to be largely composed of single isomeric fengycin species with m/z 1449.8, 1463.8, 1477.8, and 1505.8, respectively. The representative ESI-MS/MS spectra in Figure 2 illustrate the distribution of key product ions (y 8 and y 9 ) representing the linear N-terminus and the cyclic C-terminus segments of diverse fengycin species. Figure 2a shows the ESI-MS/MS spectrum of the fengycin ion at m/z 1463.8 yielding two intense product ions at m/z 966.5 and 1080.5 representing fengycin A, while the ESI-MS/MS spectrum of a fengycin ion at m/z 1505.8 (Figure 2b) yielded key product ions at m/z 994.4 and 1108.5, which readily identify it as a fengycin B homologue. Based on these key product ions, for these examples, two classes of fengycins; fengycin A ([M + H]+: 1435.8, 1449.8, 1463.8, 1477.8, 1491.8, and 1505.8) and fengycin B ([M + H]+ : 1463.8, 1477.8, 1491.8, 1505.8, 1519.8, 1533.8, and 1547.8) (see Supplemental Table S1) are identified.
Considering the results of mass spectrometric analysis, one caveat may be noted. In the present study, specific residues have been inferred at the individual residue positions based on the mass shifts observed upon fragmentation. For example, a shift of 71 Da has been assigned to an Ala while a shift of 129 Da is assigned to a Glu residue. In principle, assignments based exclusively on the basis of mass spectrometry may not readily distinguish between isomeric residues like N-methyl Gly71 and Ala71, N-methyl Asp129 and Glu129, N-methyl Val113 and Ile/Leu113. Bacterial peptides biosynthesized by NRPSs can indeed incorporate wide residue variations. However, the assignments reported in the present studies are based on the extensive literature of fengycins. Independent characterization by other techniques like NMR spectroscopy or amino acid analysis is rendered difficult by the extensive microheterogeneity of this natural mixture, with multiple overlapping components in each HPLC fractions. Indeed, mass spectrometry posses an unusual strength in being able to characterize these multiple component mixtures.
3.3 New Fengycin Classes
Interestingly, Figure 2c–e show product ions that are not diagnostic of any of these two classes. For example, in Figure 2c, ESI-MS/MS of the ion at m/z 1449.8 shows key product ions at m/z 952.4 (y 8 ) and 1066.5 (y 9 ); Figure 2d shows the presence of m/z 980.5 and 1094.5 as diagnostic product ions. Figure 2e shows the product ions y 8 , y' 8 , y 8 , and y' 9 obtained from the fragmentation of fengycin molecule with m/z 1519.8 from peak P19. The observed product ions in the MS/MS spectrum of fengycin at 1519.8 provide evidence for the presence of two isomeric fengycins [fengycin B (994.4 and 1108.5) and new fengycin (1008.4 and 1122.5)]. Surprisingly, the MS/MS spectrum of 1519.8 shows an unusual intensity distribution in the isotopic multiplets of the precursor ion (1519.8) as well as the product ions (1008, 1022). The second isotopic peak in each multiplet is significantly more intense than anticipated from the molecular formula. This feature will be discussed in a subsequent section.
In order to further characterize the fengycin mixture produced by Bacillus subtilis K1, the isolated HPLC fractions were subjected to mild alkaline hydrolysis to open the macrocyclic lactone ring, facilitating mass spectrometric sequencing of the derived linear molecules [24, 36].
3.4 Fengycin A2 (Ala6, Val10) and Fengycin B2 (Val6, Val10)
Further sequence assignment of the fengycin ion at m/z 1449.8, yielding two intense product ions at m/z 952.4 and 1066.5 (Figure 2c), is carried out using the linearized product obtained after saponification (m/z 1467.8) (Figure 3a). Opening of the lactone ring is expected to result in a mass increase of 18 Da. Figure 3a shows an ESI-MS/MS spectrum in which product ions of both b- and y-series are clearly identifiable, permitting unambiguous sequence assignment. As seen from the assignment in Figure 3a, the fengycin eluting in peak P13 contains Ala at position 6 and Val at position 10. In the literature, fengycins have been classified into two classes, A and B, corresponding to the presence of DAla /DVal at position 6. In all fengycins characterized thus far, Ile is present at position 10. Thus, the peak P8 with m/z 1449.8 obtained in the B. subtilis K1 extract contains a new variant of fengycin A termed as fengycin A2, with an amino acid replacement of Ile by Val at position 10. MS/MS analysis of the intact protonated precursor ions at m/z 1421.8, 1435.8, and 1463.8 also yielded the same intense product ions as observed in the MS/MS spectrum of 1449.8, which are also assigned as new macrocyclic fengycin A2 variants (Ala6 and Val10) with a variation in the chain length of the β-OH FA acid. C14, C15, and C17, fatty acids are consistent with the data for m/z 1421.8 1435.8, 1463.8, respectively (Scheme 1b) (see Supplemental Table S1).
Similarly, Figure 3b shows the MS/MS spectrum of the linearized precursor of an intact fengycin at m/z 1477.8 (Figure 2d, product ions 980.5 and 1094.5), with an m/z value of 1495.8. The observed b- and y-fragment ions permitted coverage of the entire sequence, establishing that the fengycin with m/z 1477.8 corresponds to a new variant of fengycin B (termed as fengycin B2) that contains Val residues at both 6 and 10 positions (Figure 3b). The MS/MS of intact protonated ions at m/z 1449.8, 1463.8, 1491.8, 1505.8, 1519.8, 1533.8, and 1547.8 also yielded the same intense paired diagnostic product ions, as observed in case of 1477.8. These are also assigned to new fengycin B2 homologues (Val6 and Val10), with a variation in chain length of the β-OH FA (C14–C21) (see Supplemental Table S1) and (Scheme 1b). The occurrence of fengycin variants with Val at position 10 has also been previously suggested from a study of lipopeptides produced by a Bacillus megatarium strain [12].
3.5 Fengycin C (Leu/Ile6, Ile10)
The ESI-MS/MS fragmentation of the ion with an m/z 1519.8, obtained from HPLC peak P19, presents an interesting example of a situation where two isomeric species coelute. The ESI-MS/MS spectrum shown in Figure 2e establishes the presence of two distinct fengycins, with the fragment ion pairs 994.4/1008.4 and 1108.5/1122.5. A linearized product at m/z 1537.9, obtained following saponification, was subjected to further ESI-MS/MS fragmentation. As shown in Figure 3c, the observed fragment ions could be assigned to the presence of two distinct sequences, which differ in both the β-OH FA residue and at position 6 in the macrocycle. A Leu/Ile to Val replacement at position 6 together with an increase in the β-OH FA chain length from C17 to C18 results in two distinct molecules of identical mass. Thus, the product ion pair 1008.5 and 1122.5 corresponds to fengycin C, a third new variant of fengycin (Scheme 1b). The MS/MS of the cyclic protonated precursor ions at m/z 1489.8, 1505.8, 1533.8, and 1547.8 also yielded same intense product ion pairs as observed in the MS/MS spectrum of m/z 1519.8. These species are assigned as new fengycin C variants (Ile/Leu6 and Leu10), with a variation in the chain length of the β-OH FA corresponding to C15–C19 chains (see Supplemental Table S1).
3.6 Fengycins with Unsaturated β-Hydroxy Fatty Acid
The difficulties encountered in the analysis of extremely micro-heterogeneous peptides derived from microbial sources may be best illustrated by consideration of HPLC peak P19, whose MALDI-MS spectrum is shown in Figure 4. At least, eleven distinct masses (m/z 1433.9, 1447.9, 1462.0, 1476.0, 1478.0, 1490.0, 1492.0, 1506.0, 1520.0, 1534.0, and 1547.9) are identifiable. The presence of sodium adducts (M + Na)+ further enhances the complexity of the observed spectrum. The most intense ion observed had m/z value 1447.9. Interestingly, the fengycins characterized in this mass range thus far from B. subtilis K1 extract correspond to masses 1421.8, 1435.8, 1449.8, 1463.8, 1477.8, 1491.8, 1505.8, 1519.8, 1533.8, and 1547.8. The ion with an m/z 1447.9 corresponds to a mass (i.e., 2 Da lower than the species with mass 1449.9. This may be ascribed to the presence of a double bond in the β-OH FA moiety, a structural feature that has been previously characterized [11, 19, 25]. Further confirmation is obtained by the analysis of ESI-MS/MS spectra of both the cyclic species and the linearized product following saponification, as shown in Supplemental Figures S1a, b. The cyclic molecule fragments to yield m/z ions 966.5 and 1080.5, corresponding to the macrocyclic moiety and the macrocycle along with the Orn residue, respectively (see Supplemental Figure S1a). Both these m/z ions match those previously obtained for fragment ions of fengycin A (Scheme 1a). Thus, the reduction of 2 Da must be assigned to the N-terminus β-OH FA. Further confirmation is obtained by MS/MS analysis of the linearized molecule following saponification (see Supplemental Figure S1b). The strain of b- and y-ions suggests the presence of a C15 chain, which contains the site of unsaturation. Previous studies of fengycin have established unsaturation at the C13–C14 bond in the C15 β-OH FA [25]. Careful inspection of Figure 4 reveals parent ions with m/z values 1433.9, 1447.9, 1462.0, 1476.0, and 1490.0, all of which show a reduction in mass of 2 Da from the fengycins characterized earlier, suggesting they have also arisen from the incorporation of C15 β-OH FA. The ions at m/z 1433.8, 1447.8, 1461.8, 1475.8, and 1489.8 were identified by ESI-MS/MS analysis. The peptide sequences of these monounsaturated C15 β-OH FA fengycins are summarized in Supplemental Table S1.
3.7 Gln/Glu Variability and Microheterogeneity of Fengycins
The proceeding section emphasized the complexity of the crude mixture of the coeluting fengycins. Evidence of microheterogeneity is also apparent when the intensities of various components of an isotopic multiplet are examined. This feature is exemplified in the ESI-MS of HPLC peak P14 (Figure 5a), which exhibits a cluster of fengycins in the mass range m/z 1449–1510. Figure 5a inset shows the intensity of the isotopic components corresponding to the multiplet with m/z 1449.8. It is observed that the component with m/z 1450.8 is appreciably more intense than the m/z 1449.8 peak, suggesting the presence of two molecular species differing in mass by 1 Da. The calculated intensity distribution (Isotope Distribution calculator, IDCalc, ver. 0.3, [37]) of the isotopic multiplet using a molecular formula C71H108O20N12 is shown in the inset of Figure 5a. The computed isotopic distribution for the multiplet presented in Figure 5a suggests that the observed distribution does indeed arise from an overlap of mass spectral peaks from two fengycin species, differing in mass by 1 Da. The second species with an increase in mass of 1 Da may be attributed to a fengycin species in which a Glu residue (m/z 1450.8) replaces a Gln residue (m/z 1449.8). Inspection of the components of peak P14 at m/z 1463.8, 1477.8, and 1491.8 suggests that in all cases, two distinct species differing in 1 Da are present. Thus far, all reported fengycins contain Glu residues at positions 1 and 5 and a Gln residue at position 8 (Scheme 1a). The results described above suggest that the Bacillus subtilis K1 lipopeptide extract does indeed contain fengycin species, which posses three Glu residues. The presence of fengycin species with two or three Glu residues are further confirmed by esterification of the free carboxylic acid residues of each fengycin species of HPLC peak P14, using a benzyl alcohol-acetyl chloride mixture (50:1 vol/vol). Under these conditions, benzyl esters are formed, resulting in a mass increase (ΔM) of 90 Da. Esterification using methanol was initially attempted, resulting in a mass increase of 14 Da upon methyl ester formation, which resulted in m/z values identical to those obtained for the fengycin components differing by 14 Da or integral multiples thereof (fatty acid variants and Ile/Val or Ile/Leu/Val/Ala replacements). Supplemental Figure S2a shows the mass spectrum of the HPLC peak 14 after esterification with benzyl alcohol for 30 min. The two clusters at m/z 1630.0 to 1695.0 and 1730 to 1757 correspond to species containing 2Glu (ΔM = 180) and 3Glu (ΔM = 270) benzyl ester functionalities. This is clearly indicative of the presence of two distinct sets of fengycin species, one of which contains two Glu1,5 and one Gln8 residues, whereas the other contains 3Glu1,5,8 residues. The insets in Supplemental Figure S2a show the observed and theoretically computed intensities of the isotopic multiplets for the dibenzyl and tribenzyl esters. In both the cases, there is a good agreement between calculated and observed distribution of intensities. One question that may arise in interpreting the chemical derivatization results is whether the third Glu residue arises because of the acid catalyzed hydrolysis of the Gln side chain under the esterification conditions. In order to address this question, two sets of control experiments were performed. Iturin A in HPLC peak P4 (see Supplemental Figure S3, Table S1), which contains 1Gln4 and 3Asn1,3,6 residues and lacks any Glu/Asp residue was subjected to identical esterification conditions. The absence of any detectable hydrolysis and subsequent esterification of iturins suggests that carboxamide hydrolysis is a slow process and may be insignificant at the short reaction times used. Supplemental Figure S3 provides evidence for small amounts of hydrolysis upon prolonged exposure to the benzyl alcohol-acetyl chloride mixture. In a second set of experiments, HPLC peak P17 containing a fengycin cluster with m/z 1478.1 to 1531.3 was chosen for esterification. The most intense component at m/z 1492.1 shows an isotopic multiplet which suggests a single fengycin species with 2 Glu and 1 Gln residues (see Supplemental Figure S4, control). Exposure to esterification condition over a time period of 1–3 h yielded only dibenzyl esters, confirming the absence of the Gln to Glu conversion under these conditions. Significantly longer reaction periods of ~10 h are needed for the detection of tribenzyl esters (see Supplemental Figure S4). Interestingly, the triesters show an additional mass increase of 18 Da, corresponding to acid catalyzed lactone ring opening (see Supplemental Figure S4). The results of the esterification of P14 and the two control experiments described above provide strong supporting evidence for the presence of fengycin species containing three Glu residues, at positions 1, 5, and 8, in the B. subtilis K1 lipopeptide extract.
ESI-MS spectra of HPLC peaks (a) P14, (b) P19, and (c) P6 (masses are indicated as m/z values for the first component of the cluster), acquired in a Q-TOF mass spectrometer. (Insets) Observed and calculated isotope distributions of representative ion clusters at m/z 1449.8 (P14) (a), 1489.8 (P19) (b), and 1467.5 (P6). The isotopic distributions are calculated using Isotope Distribution calculator [33]
At this point, it became necessary to establish that acidic conditions during the extraction of the lipopeptide mixture did not result in artifactual hydrolysis. The lipopeptide extract was obtained by precipitation from a culture supernatant that had been adjusted to pH 2 and stored at 4 °C for 2 h. HPLC peak 17, which contains fengycins with 2 Glu and 1 Gln residues, was examined after incubation at pH 2 for a time period extending up to 15 h. No change was observed in the major components with m/z 1478.1 and 1492.1, suggesting that the Gln residue remains intact under these conditions (see Supplemental Figure S5).
Further examples of additional microheterogeneity may be observed in the cluster of fengycin ions as shown in Figure 5b. The inset illustrates a representative example of the isotopic distribution observed for a cluster with the lowest m/z value of 1489.8. The sequence of fengycin with m/z 1489.8 has been previously assigned (see Supplemental Table S1), with the molecular formula C74H112O20N12. The isotopic pattern computed for this component shown in Figure 5b (inset) clearly establishes that this observed distribution of intensities is due to heterogeneity. Inspection of the intensity pattern suggests the presence of at least three distinct species with m/z 1489.8, 1491.8, and 1492.8. The additional species, which yield mass increase of 1 Da and 2 Da, respectively, may be assigned to a species with Glu substituting for Gln at position 8 and a saturated C15 β-OH FA to unsaturated C15 β-OH FA change at the N-terminus. The anticipated isotopic distribution for the fengycin ion with m/z 1489.8 is shown in Figure 5b inset. Thus, the observed distribution of intensities for the 1489.8–1492.8 ion cluster may be explained by the presence of three distinct fengycin species. Further confirmation of this assignment is obtained by esterification with a benzyl alcohol-acetyl chloride mixture, which demonstrates the species with both di- and tribenzyl esters (see Supplemental Figure S2b). Inspection of the isotopic multiplets obtained for the diester (m/z 1669.9–1676.0) and triester (m/z 1762.9–1766.0) suggest that the species containing three Glu residues posses a saturated β-OH FA chain. In contrast, species that posses two Glu and one Gln residues appear to have both unsaturated and saturated β-OH FAs.
Further confirmation of the fengycin species differing by 1 Da (due to variation in Glu/Gln) is obtained by high resolution mass spectrometry. In order to determine the 1 Da variation, an isotopic multiplet with m/z 1477.8 is further selected for the MS/MS analysis. The ESI-MS/MS spectrum of a fengycin species with an m/z 1477.8 shows the presence of two product ions with m/z 980.5 and 1094.5 (Figure 6a). Interestingly, the product ions also show an abnormal intensity distribution within the isotopic multiplets. The sequence of the m/z 1477.8 ion is determined by fragmenting the corresponding ion in the linearized molecule (m/z 1495.8) (Supplemental Table S1). Figure 6b shows the isotopic distribution of all the observed as well as calculated b- and b'-ions observed in the MS/MS spectra of m/z1495.8 (a linearized form of cyclic fengycin ion with m/z 1477.8). Careful inspection of the ion at m/z 1495.8 in the linearized fengycin shows a normal intensity distribution in the multiplets of the b 1 –b 7 product ions. In sharp contrast, the intensity distribution in the b 8 –b 9 ion multiplets is abnormal, resembling that observed for the cyclic species (m/z 1477.8) and the product ions (m/z 980.5 and 1094.5). This clearly suggests the presence of two fengycin species differing by 1 Da as a result of the Glu to Gln change. In the spectrum of the linearized species, the b' 1 –b' 7 fragment ions have same mass as the b 1 –b 7 ions for the both fengycin species whereas, the b' 8 and b' 9 (Glu8) ions are 1 Da more than the b 8 and b 9 . This confirms the variation of Glu/Gln at position 8. Inspection of the remaining multiplets of peak P14 at m/z 1449.8, 1463.8, and 1491.8 also suggests that in all cases, two distinct fengycin species (2 Glu + 1 Gln and 3 Glu) differing by 1 Da are present. The results described above suggest that the Bacillus subtilis K1 lipopeptide extract does indeed contain species that posses Glu residues at all three positions (1, 5, and 8). In this study, mass spectrometry facilitates the identification of new fengycin species differing by 1 Da due to the variation of Gln/Glu at position 8. The Gln8/Glu8 variants of fengycins identified in the extract of B. subtilis K1 are listed in Supplemental Table S1.
(a) ESI-MS/MS spectrum of a protonated cyclic fengycin ion at m/z 1477.8 and (b) observed and calculated intensity distributions of b-/b'- product ion multiplets of a linearized protonated fengycin ion (of corresponding cyclic fengycin ion at m/z 1477.8) at m/z 1495.8. Both the cyclic and linearized fengycin MS/MS spectra are acquired in a Q-TOF mass spectrometer
Mass spectral analysis of HPLC peak 6 (Figure 5c) reveals the presence of linearized fengycin molecules, which may be precursors for the cyclization reaction, the last step of the biosynthetic process [38]. Figure 5c shows the presence of m/z ions at 1453.5, 1467.5, 1481.5, and 1495.5, all of which correspond to the linear species derived by the opening of the lactone ring of fengycins with m/z 1435.8, 1449.8, 1463.8, and 1477.8. Lactone hydrolysis during processing of the B. subtilis K1 extract may be ruled out by exposing the cyclic fengycins to conditions identical to that used in the initial extraction (pH 2, incubation at 4 °C for 2 h). No detectable hydrolysis could be demonstrated under these conditions (see Supplemental Figure S5). The detection of biosynthetic precursors in the culture supernatant may be anticipated. Inspection of the isotopic distribution of an ion cluster with m/z 1467.5 to 1471.5 suggests that linear species that contain 3 Glu residues are dominant over a species containing 2 Glu and 1 Gln residues (Figure 5c inset). Linear fengycins could also be detected, albeit in very small quantities, by direct MALDI-MS analysis of cell suspensions, without exposure to acidic condition (see Supplemental Figure S6). The identity of this linear species was further established by the fragmentation patterns observed in MALDI-MS/MS of the m/z 1467.5 spectrum (see Supplemental Figure S7). The observed train of b- and y-ions confirms the sequence.
4 Conclusions
The present study demonstrates the presence of lipopeptides belonging to the surfactin, iturin, and fengycin families in the extract of the banyan endophyte, B. subtilis K1, which exhibits significant antifungal activity against 11 test fungi [35]. The mass spectrometric analysis of fengycin described in this study permits the identification of several new fengycin variants. Three sites of variation are readily demonstrated: (1) homologues of β-OH FA, (2) variations at position 6 (Ala/Val/Ile/Leu), and (3) variations at position 10 (Val/Ile). In addition, the presence of unsaturated fatty acids is suggested by the 2 Da reduction in mass of some fengycins. Previous studies have reported fengycins with unsaturated β-OH FAs of C15 to C17 chain length [13, 21, 25], whereas in our case, fengycins with unsaturated fatty acids correspond to C15 chains. The observation of anomalous intensities of isotopic patterns in the mass spectra of fengycin fractions has allowed the characterization of new variants, Glu8 in place of Gln8. A convenient esterification protocol using benzyl alcohol permits a facile distinction between fengycin species containing 2 Glu and 3 Glu residues. Analysis of isotope multiplet intensities in the fragment ions further confirms the identity of fengycin species differing by 1 Da by localizing the variable position (Gln/Glu8) in the fengycin variants. The extensive microheterogeneity appears to be generated by reduced fidelity of the biosynthesis at four sites, namely, the β-OH FA and residues at position 6, 8, and 10 in the macrocyclic ring. The remarkable heterogeneity and complexity of the lipopeptides produced by B. subtilis and related organisms have always raised questions of the utility of the molecules to the producing organisms [14]. Clearly, their antimicrobial properties suggest that lipopeptide production may indeed confer an advantage for survival in an environment in which other organisms compete for nutrients [6, 13, 14]. The lack of fidelity at specific sites during the process of lipopeptide assembly on the non-ribosomal peptide synthetases [38] suggests that no evolutionary advantage is achieved by making the requirements for residue incorporation more stringent at these sites. Whether microheterogeneity confers a selective advantage remains to be established.
References
Strobel, G., Daisy, B.: Bioprospecting for microbial endophytes and their natural products. Micorbiol. Mol. Biol. Rev. 67, 491–502 (2003)
Zhang, H.W., Song, Y.C., Tan, R.X.: Biology and chemistry of endophytes. Nat. Prod. Rep. 23, 753–771 (2006)
Khare, C.P.: Indian Medicinal Plants, An Illustrated Dictionary, p. 264. Springer Reference, New York (2007)
Sieber, T.: Endophytic fungi of forest trees: Are they mutualists? Fungal Biol. Rev. 21, 75–89 (2007)
Suryanarayanan, T.S., Vijaykrishna, D.: Fungal endophytes of aerial roots of Ficus benghalensis. Fungal Diversity. 8, 155–161 (2001)
Stein, T.: Bacillus subtilis antibiotics: Structures, synthesis and specific functions. Mol. Microbiol. 56, 845–847 (2005)
Ng, J., Bandeira, N., Liu, W.T., Ghassemian, M., Simmons, T.L., Gerwick, W.H., Linington, R., Dorrestein, P.C., Pevzner, P.A.: Dereplication and De Novo sequencing of nonribosomal peptides. Nat. Methods 6, 596–599 (2009)
Caboche, S., Leclere, V., Pupin, M., Kuchrov, G., Jacques, P.: Diversity of monomers in nonribosomal peptides: Towards the prediction of origin and biological activity. J. Bacteriol. 192, 5143–5150 (2010)
Aric, A., Jacques, P., Wathelet, B., Paquot, M., Fuchs, R., Budzikiewics, H., Thonart, P.: Influence of culture conditions on lipopeptide production by Bacillus subtilis. Appl. Biochem. Biotechnol. 91/93, 1–9 (2001)
Yu, G.Y., Sinclair, J.B., Hartman, G.L., Bertagnolli, B.L.: Production of Iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol. Biochem. 34, 955–963 (2002)
Vater, J., Kablitz, B., Wilde, C., Franke, P., Mehta, N., Cameotra, S.S.: Matrix-assisted laser desorption ionization-time of flight mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of Bacillus subtilis C-1 isolated from petroleum sludge. Appl. Environ. Microbiol. 68, 6210–6219 (2002)
Pueyo, M.T., Bloch Jr., C., Carmon-Ribeiro, A.M., di Mascio, P.: Lipopeptides produced by a soil Bacillus megaterium Strain. Microb. Ecol. 57, 367–378 (2005)
Nagorska, K., Bikowski, M., Obuchowski, M.: Multicellular behaviour and production of a wide variety of toxic substances support usage of Bacillus subtilis as a powerful biocontrol agent. Acta Biochim. Pol. 54, 495–508 (2007)
Ongena, M., Jacques, P.: Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125 (2008)
Arguelles-Aria, A., Ongena, M., Halimi, B., Lara, Y., Brans, A., Joris, B., Fickers, P.: Microb.: Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Fact. 26, 1–12 (2009)
Arima, K., Kakinuma, A., Tamura, G.: Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 31, 88–494 (1968)
Garbay-Jaureguiberry, C., Roques, B.P., Delcambe, L., Peypoux, F., Michel, G.: NMR conformational study of Iturin A, an antibiotic from Bacillus subtilis. FEBS Lett. 93, 151–156 (1978)
Winkelmann, G., Allgaier, H., Lupp, R., Jung, G.: Iturin AL-A new long chain iturin a possessing an unusual high content of C16-beta-amino acids. J. Antibiot. 36, 1451–1457 (1983)
Vanittanakom, N., Loeffler, W., Koch, U., Jung, G.: Fengycin-A novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot. 39, 888–901 (1986)
Kowall, M., Vater, J., Kluge, T., Stein, P., Ziessow, D.: Separation and characterization of surfactin isoforms produced by Bacillus subtilis OKB 105. J. Colloid Interface Sci. 204, 1–8 (1998)
Volpon, L., Besson, F., Lancelin, J.M.: NMR structure of antibiotics plipastatins A and B from Bacillus subtilis inhibitors of phospholipase A(2). FEBS Lett. 485, 76–80 (2000)
Madonna, J., Voorhees, K.J., Taranenko, N.I., Laiko, V.V., Doroshenko, V.M.: Detection of cyclic lipopeptide biomarkers from Bacillus species using atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 75, 1628–1637 (2003)
Pecci, Y., Rivardo, F., Martinotti, M.G., Allegrone, G.: LC/ESI-MS/MS characterisation of lipopeptide biosurfactants produced by the Bacillus licheniformis V9T14 strain. J. Mass Spectrom. 45, 772–778 (2010)
Williams, B.H., Hathout, Y., Fenselau, C.: Structural characterization of lipopeptide biomarkers isolated from Bacillus globigii. J. Mass Spectrom. 37, 259–264 (2002)
Kim, P.I., Bai, H., Bai, D., Chae, H., Chung, S., Kim, Y., Park, R., Chi, Y.T.: Purification and characterization of a lipopeptide produced by Bacillus thuringiensis CMB26. J. Appl. Microbiol. 97, 942–949 (2004)
Nihorimbere, V., Cawoy, H., Seyer, A., Brunelle, A., Thonart, P., Ongena, M.: Impact of rhizosphere factors on cyclic lipopeptide signature from the plant beneficial strain Bacillus amyloliquefaciensS499. FEMS Microbiol. Ecol. 79, 176–191 (2012)
Nishikiori, T., Naganawa, H., Muraoka, Y., Aoyagi, T., Umezawa, H.: Plipastatins: New inhibitors of phospholipase A2, produced by Bacillus cereus BMG302-fF67. III. Structural elucidation of plipastatins. J. Antibiot. 39, 755–761 (1986)
Peypoux, F., Besson, F., Michel, G., Delcambe, L.: Structure of bacillomycin D, a new antibiotic of the Iturin group. Eur. J. Biochem. 118, 323–327 (1981)
Peypoux, F., Pommier, M.T., Das, B.C., Besson, F., Delcambe, L., Michel, G.: Structures of bacillomycin D and bacillomycin L peptidolipid antibiotics from Bacillus subtilis. J. Antibiot. 37, 1600–1604 (1984)
Peypoux, F., Pommier, M.T., Marion, D., Ptak, M., Das, B.C., Michel, G.: Revised structure of mycosubtilin, a peptidolipid antibiotic from Bacillus subtilis. J. Antibiot. 39, 636–641 (1986)
Grangemard, I., Wallach, J., Maget-Dana, R., Peypoux, F.: Lichenysin: A more efficient cation chelator than surfactin. Appl. Biochem. Biotechnol. 90, 199–210 (2001)
Naruse, N., Tenmyo, O., Kobaru, S., Kamei, H., Miyaki, T., Konishi, M., Oki, T.: Pumilacidin, a complex of new antiviral antibiotics. Production, isolation, chemical properties, structure and biological activity. J. Antibiot. 43, 267–280 (1990)
Isogai, A., Takayama, S., Murakoshi, S., Suzuki, A.: Structure of β-amino acids in antibiotics Iturin A. Tetrahedron Lett. 23, 3065–3068 (1982)
Gong, M., Wang, J.D., Zhang, J., Yang, H., Lu, X.F., Pei, Y., Cheng, J.Q.: Study of the antifungal ability of Bacillus subtilis strain PY-1 in vitro and identification of its antifungal substance (iturin A). Acta Biochim. Biophys. Sin. (Shanghai) 38, 233–240 (2006)
Pathak, K.V.: Purification and Characterization of Antifungal Compounds Produced by Banyan Endophytic Bacilli. Ph.D. Thesis, Sardar Patel University, Vallabh Vidynagar, India, p 42 (2011)
Sabareesh, V., Ranganayaki, R.S., Raghothama, S., Bopanna, M.P., Balaram, H., Srinivasan, M.C., Balaram, P.: Identification and characterization of a library of microheterogeneous cyclohexadepsipeptides from the fungus isaria. J. Nat. Prod. 70, 715–729 (2007)
Kubinyi, H.: Calculation of isotope distributions in mass spectrometry. A trivial solution for a non-trivial problem. Anal. Chim. Acta 247, 107–119 (1991)
Sieber, S.A., Marahiel, M.A.: Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chem. Rev. 105, 715–738 (2005)
Acknowledgments
K.P. and H.K. acknowledge the University Grant Commission (UGC), Government of India for the financial support. K.G. acknowledges the receipt of Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), Government of India. The mass spectrometric facility was supported by a proteomics program grant from the Department of Biotechnology (DBT), Government of India. K.P. thanks Sunita Prakash and Sonia Kulkarni, for assisting with the mass spectrometers. Authors also thank Professor Hema Balaram, Jawaharlal Nehru Center for Advanced Scientific Research, Bangalore for the use of the MALDI-TOF mass spectrometer.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1.55 MB)
Rights and permissions
About this article
Cite this article
Pathak, K.V., Keharia, H., Gupta, K. et al. Lipopeptides from the Banyan Endophyte, Bacillus subtilis K1: Mass Spectrometric Characterization of a Library of Fengycins. J. Am. Soc. Mass Spectrom. 23, 1716–1728 (2012). https://doi.org/10.1007/s13361-012-0437-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-012-0437-4