Abstract
Pigeonpea production is severely constrained by wilt disease caused by Fusarium udum. In the current study, we discover the putative genomic regions that control resistance response to variant 2 of fusarium wilt using association mapping approach. The association panel comprised of 89 diverse pigeonpea genotypes including seven varieties, three landraces and 79 germplasm lines. The panel was screened rigorously for 3 consecutive years (2013–14, 2014–15 and 2015–2016) against variant 2 in a wilt-sick field. A total of 65 pigeonpea specific hypervariable SSR markers (HASSRs) were screened representing seven linkage groups and 29 scaffolds of the pigeonpea genome. A total of 181 alleles were detected, with average values of gene diversity and polymorphism information content (PIC) of 0.55 and 0.47, respectively. Further analysis using model based (STRUCTURE) and distance based (clustering) approaches separated the entire pigeonpea collection into two distinct subgroups (K = 2). The marker trait associations (MTAs) were established based on three-year wilt incidence data and SSR dataset using a unified mixed linear model. Consequently, six SSR markers were identified, which were significantly associated with wilt resistance and explained up to 6% phenotypic variance (PV) across the years. Among these SSRs, HASSR18 was found to be the most stable and significant, accounting for 5–6% PV across the years. To the best of our knowledge, this is the first report of identification of favourable alleles for resistance to variant 2 of Fusarium udum in pigeonpea using association mapping. The SSR markers identified here will greatly facilitate marker assisted resistance breeding against fusarium wilt in pigeonpea.





Similar content being viewed by others
References
Agbagwa IO, Datta S, Patil PG, Singh P, Nadarajan N (2012) A protocol for high-quality genomic DNA extraction from legumes. Genet Mol Res 11:4632–4639
Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y et al (2010) Genome wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465:627–631
Bohra A (2013) Emerging paradigms in genomics-based crop improvement. Sci World J 2013:585467
Bohra A, Dubey A, Saxena RK, Penmetsa RV, Poornima KN, Kumar N et al (2011) Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea. BMC Plant Biol 11:56
Bohra A, Jha R, Pandey G, Patil PG, Saxena RK, Singh IP et al (2017) Genome wide hypervariable SSR markers for molecular breeding applications in pigeonpea [Cajanus cajan (L.) Millspaugh]. Front. Plant Sci 8:377
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633
Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172:1165–1177
Butler EJ (1906) The wilt disease of pigeonpea and pepper. Agric J India 1:25–26
Dhar V (1995) Disease resistant lines with stable resistance identified as donor parents under AICPIP – Pathology programme from 1984–1994. Consolidated report of kharif pulses, plant pathology. All India co-ordinated pulses improvement project (ICAR). Kanpur, IIPR, P.IV
Dubey A, Farmer A, Schlueter J, Cannon SB, Abernathy B, Tuteja R et al (2011) Defining the transcriptome assembly and its use for genome dynamics and transcriptome profiling studies in pigeonpea (Cajanus cajan L.) DNA Res 18:153–164
Dutta S, Kumawat G, Singh BP, Gupta DK, Singh S, Dogra V et al (2011) Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol 11:17
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
FAOSTAT (2014) Food and agriculture organization of United Nations. http://faostat.fao.org
Gomez KA, Gomez AA (1984) Statistical procedure for agricultural research, 2nd edn. Wiley, NewYork
Gupta PK, Rustgi S, Kulwal PL (2005) Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol 57:461–485
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113:163–185
Hu Z, Zhang D, Zhang G, Kan G, Hong D, Yu D (2014) Association mapping of yield-related traits and SSR markers in wild soybean (Glycine soja Sieb. and Zucc.) Breed Sci 63:441–449
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y et al (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961–967
Jain KC, Reddy MV (1995) Inheritance of resistance to fusarium wilt in pigeonpea (Cajanus cajan L.) Indian J Genet Plant Breed 55:434–443
Joshi AR (1957) Genetics of resistance to disease and pests. Indian J Genet Plant Breed 17:305–317
Kannaiyan J, Nene YL (1981) Influence of wilt at different growth stages on yield loss in pigeonpea. Trop Pest Manage 27:141
Kassa MT, Penmetsa RV, Carrasquilla-Garcia N, Sarma BK, Datta S, Upadhyaya HD et al (2012) Genetic patterns of domestication in pigeonpea (Cajanus cajan (L.) Millsp.) and wild Cajanus relatives. PLoS One 7:e39563. doi:10.1371/journal.pone.0039563
Kotasthane SR, Gupta O (1981) Trend of wilting in pigeonpea varieties of variable maturity. Abstracts of papers of the third symposium of plant pathology, Indian Phytopathological Society, 14–18 Dec 1981. New Delhi, India, p 161–162
Kotresh H, Fakrudin B, Punnuri SM, Rajkumar BK, Thudi M, Paramesh H et al (2006) Identification of two RAPD markers genetically linked to a recessive allele of a fusarium wilt resistance gene in pigeonpea (Cajanus cajan L. Millsp.) Euphytica 149:113–120
Kudapa H, Bharti AK, Cannon SB, Farmer AD, Mulaosmanovic B, Kramer R et al (2012) A comprehensive transcriptome assembly of pigeonpea (Cajanus cajan L.) using Sanger and second-generation sequencing platforms. Mol Plant 5:1020–1028. doi:10.1093/mp/ssr111
Kumar Y, Kwon SJ, Coyne CJ, Hu J, Grusak MA, Kisha TJ et al (2014) Target region amplification polymorphism (TRAP) for assessing genetic diversity and marker trait associations in chickpea (Cicer arietinum L.) germplasm. Genet Resour Crop Evol 61:965–977
Kumawat G, Raje RS, Bhutani S, Pal JK, Mithra ASVCR, Gaikwad K et al (2012) Molecular mapping of QTLs for plant type and earliness traits in pigeonpea (Cajanus cajan L. Millsp.) BMC Genet 13:84
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Mir RR, Kudapa H, Srikanth S, Saxena RK, Sharma A, Azam S (2014) Candidate gene analysis for determinacy in pigeonpea (Cajanus spp.) Theor Appl Genet 127:2663–2678
Mir RR, Saxena RK, Saxena KB, Upadhyaya HD, Kilian A, Cook DR et al (2012) Whole-genome scanning for mapping determinacy in pigeonpea (Cajanus spp.) Plant Breed 132:472–478
Mishra S, Dhar V (2003) Variability in isolates of Fusarium udum Butler, the wilt pathogen of pigeonpea. In: Proceedings of ISMPP Zonal Conference (Eastzone). IISR, Lucknow
Mishra S, Dhar V (2010) Identification of broad base and stable sources of resistance to fusarium wilt in pigeonpea. Indian Phytopathol 63(2):165–167
Mundkur BB (1935) Influence of temperature and maturity on the incidence of sun hemp and pigeonpea wilt at Pusa. Indian J Agric Sci 5:606–618
Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE et al (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21:2194–2202
Nadimpalli RG, Jarret RL, Phatak SC, Kochart G (1994) Phylogenetic relationships of the pigeonpea (Cajanus cajan) based on nuclear restriction fragment length polymorphism. Genome 36:216–223
Natarajan M, Kannaiyan J, Willey RW, Nene YL (1985) Studies on the effects of cropping system on fusarium wilt of pigeonpea. Field Crops Res 10:333–346
Nordborg M, Weigel D (2008) Next-generation genetics in plants. Nature 456:720–723
Okiror MA (1999) Evaluation of pigeonpea (Cajanus cajan) germplasm for resistance to fusarium wilt. Indian J Agric Sci 69:600–601
Okiror MA (2002) Genetics of resistance to Fusarium udum in pigeonpea (Cajanus cajan (L.) Millsp.) Indian J Genet Plant Breed 62:218–220
Patil PG, Byregowda M, Patil BR, Das A, Mary Reena GA, Sowjanya MS et al (2016) Microsatellite markers linked to sterility mosaic disease resistance in pigeonpea (Cajanus cajan L. Millsp.) Legume Genomics Genet 7(6):1–9
Patil P, Singh IP, Dhar V, Chaudhary RG, Datta S, Chaturvedi SK et al (2013) Variant specific studies on inheritance for resistance to fusarium wilt in pigeonpea [Cajanus cajan (L.) Millsp.] Legume Res 36(6):576–581
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. Centre de cooperation internationale en recherche agronomique pour le developpement (CIRAD), Paris
Perseguini JMKC, Oblessuc PR, Rosa JRBF, Gomes KA, Chiorato AF, Carbonell SAM et al (2016) Genome-wide association studies of anthracnose and angular leaf spot resistance in common bean (Phaseolus vulgaris L.). PLoS ONE 11(3). doi: 10.1371/journal.pone.0150506
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945
Punguluri SK, Janaiah K, Govil JN, Kumar PA, Sharma PC (2006) AFLP fingerprinting in pigeonpea (Cajanus cajan (L.) Millsp) and its wild relatives. Genet Resour Crop Evol 53:423–531
Ratnaparkhe MB, Gupta VS, Ven-Murthy MR, Ranjekar PK (1995) Genetic fingerprinting of pigeonpea [Cajanus cajan (L.) Millsp.] and its wild relatives using RAPD markers. Theor Appl Genet 91:893–898
Sakiroglu M, Sherman-Broyles S, Story A, Moore KJ, Doyle JJ, Charles Brummer E (2012) Patterns of linkage disequilibrium and association mapping in diploid alfalfa (M. sativa L.) Theor Appl Genet 125:577–590
Saxena RK, Saxena KB, Kumar RV, Hoisington DA, Varshney RK (2010) Simple sequence repeat-based diversity in elite pigeonpea genotypes for developing mapping populations to map resistance to fusarium wilt and sterility mosaic disease. Plant Breed 129:135–241
Sharma M, Ghosh R, Telangre R, Rathore A, Saifulla M, Mahalinga DM et al (2016) Environmental influences on pigeonpea Fusarium udum interactions and stability of genotypes to fusarium wilt. Front Plant Sci 7:253. doi:10.3389/fpls.2016.00253
Sharma M, Rathore A, Mangala UN, Ghosh R, Sharma S, Upadhyay HD et al (2012) New sources of resistance to fusarium wilt and sterility mosaic disease in a mini-core collection of pigeonpea germplasm. Eur J Plant Pathol 133:707–714
Shi C, Navabi A, Yu K (2011) Association mapping of common bacterial blight resistance QTL in Ontario bean breeding populations. BMC Plant Biol 11:52
Singh AP, Bhargava SN (1981) Survival studies on three species of fusarium causing wilt of pigeonpea. Phytopathol Z 100:300–311
Singh IP, Dhar V, Chaudhary RC, Pandey DK (1998) Genetic of fusarium wilt resistance in pigeonpea. In: national symposium on management of biotic and abiotic stresses in pulse crops, 26–28th June 1998. IIPR, Kanpur, p 15
Singh NK, Gupta DK, Jayaswal PK, Mahato AK, Dutta S, Singh S et al (2012) The first draft of the pigeonpea genome sequence. J Plant Biochem Biotechnol 21:98–112
Singh VK, Khan AW, Saxena RK, Kumar V, Kale SM, Sinha P et al (2016a) Next-generation sequencing for identification of candidate genes for fusarium wilt and sterility mosaic disease in pigeonpea (Cajanus cajan). Plant Biotechnol J 14:1183–1194
Singh AK, Rai VP, Chand R, Singh RP, Singh MN (2013) Genetic diversity studies and identification of SSR markers associated with fusarium wilt (Fusarium udum) resistance in cultivated pigeonpea (Cajanus cajan). J Genet 92:273–280
Singh F, Singh IP, Mazumder MD (2011) Identification of fusarium wilt-resistant sources of long-duration pigeonpea (Cajanus cajan). Indian J Agric Sci 81:1046–1051
Singh D, Sinha B, Rai VP, Singh MN, Singh DK, Kumar R et al (2016b) Genetics of fusarium wilt resistance in pigeonpea (Cajanus cajan) and efficacy of associated SSR markers. Plant Pathol J 32:95–101
Thudi M, Upadhyaya HD, Rathore A, Gaur PM, Krishnamurthy L, Roorkiwal M et al (2014) Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS One 9:e96758
Tiwari S, Dhar V (2011) Prevalence of new variants of Fusarium udum in India. Indian Phytopathol 64:243–246
Upadhyay RS, Rai B (1992) Wilt disease of pigeonpea. In: plant disease of international importance. In: Singh US, Mukhopadyaya Kumar A, Chaube HS (eds) . Prentice Hall, Engelwood cliffs, pp 388–404
Van-Berloo R (2008) Computer note: GGT 2.0: versatile software for visualization and analysis of genetic data. J Hered 99(2):232–236
Varshney RK, Paulo MJ, Grando S, Van Eeuwijk FA, Keizer LCP et al (2012) Genome-wide association analyses for drought tolerance related traits in barley (Hordeum vulgare L.) Field Crops Res 126:171–180
Varshney RK, Penmetsa RV, Dutta S, Kulwal PL, Saxena RK, Datta S et al (2010) Pigeonpea genomics initiative (PGI): an international effort to improve crop productivity of pigeonpea (Cajanus cajan L.) Mol Breed 26:393–408
Wang YH, Bible P, Loganantharaj R, Upadhyaya HD (2012) Identification of SSR markers associated with height using pool-based genome-wide association mapping in sorghum. Mol Breed 30:281–292
Yang S, Pang W, Ash G, Harper J, Carling J, Wenzl P et al (2006) Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Theor Appl Genet 113:585–595
Yang X, Yan J, Shah T, Warburton ML, Li Q, Li L et al (2010) Genetic analysis and characterization of a new maize association mapping, panel for quantitative trait loci dissection. Theor Appl Genet 121:417–431
Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA et al (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355–360
Acknowledgements
This work was financially supported by the Indian Council of Agricultural Research-Network Project on Transgenic in Crops- Sub Project- Functional Genomics in Pigeonpea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Barbara Naganowska
Electronic supplementary material
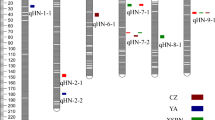
Suppl. Fig. 1
Representative partial map showing genome positions of markers associated with resistance to wilt (indicated with arrow). SSR markers were located on the reference genome using the BLAT options of genome browser in Legume information system. The marker Mb positions were converted and represented in cM map units and map was drawn with GGT software (Van-Berloo 2008) (GIF 5812 kb)
Suppl. Fig. 2
Weather graph depicting differences for temperature, relative humidity and rainfall for three cropping seasons (July–April) (Major differences were indicated by circle in the graph) (GIF 6094 kb)
Suppl. Table 1
Phenotypic data on percent wilt incidence for 89 pigeonpea genotypes for 3 years (DOC 106 kb)
Suppl. Table 2
Phenotypic effects of major marker allele at six significantly associated SSR locus against FW (DOC 36 kb)
Suppl. Table 3
Comparison of weather data for the 4 months (November to December for each year) during which phenotyping against FW was carried out (DOC 35 kb)
Rights and permissions
About this article
Cite this article
Patil, P.G., Dubey, J., Bohra, A. et al. Association mapping to discover significant marker-trait associations for resistance against fusarium wilt variant 2 in pigeonpea [Cajanus cajan (L.) Millspaugh] using SSR markers. J Appl Genetics 58, 307–319 (2017). https://doi.org/10.1007/s13353-017-0400-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-017-0400-y