Abstract
The viral agent of the porcine epidemic diarrhea (PED) was investigated during the reported 2014–2015 outbreaks in commercial farms in Central Luzon, Philippines. The study covered detection of PED virus (PEDV) in fecal and intestinal samples through reverse transcription PCR and sequence analysis of the nucleocapsid (N) gene. Results showed that 10 out of 34 fecal and intestinal samples examined were positive for PEDV. The partial nucleotide sequence of the N gene of the field samples showed 98–99% homologous to PEDV sequences registered in the GenBank. It was also noted that N gene sequences between field samples were 98% homologous. Interestingly, the partial sequences of the N genes of the field samples were genetically similar to the PEDV isolates from USA, China, Mexico, Canada and Japan. The phylogenetic tree analysis revealed that the Philippine samples clustered in group 2–1 of the PEDV, wherein the isolates of this group were responsible for the outbreaks in Asia and the USA. Analysis of the partial nucleotide and amino acid sequences revealed polymorphisms, deletions and insertions in the N-gene of the PEDV. Amino acid sequence alignment also showed deletions and insertion in the PEDV detected in the Philippines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine epidemic diarrhea (PED) is a coronaviral disease of pigs caused by the PED virus (PEDV) [1,2,3,4,5,6] which has created great alarm among hog raisers, breeders and farm owners in countries where PED outbreaks occur. PED was first recognized as a clinical entity in the UK in 1971 and has been reported as an enteric disease in growing and fattening pigs with signs similar to transmissible gastroenteritis virus (TGEV) infection [3, 4] (https://www.aphis.USDA2013.gov/animal_health/animal_dis_spec/swine/downloads/ped_tech_note.pdf). In 1978, scientists at Ghent University in Belgium identified a suspected coronaviral agent in an outbreak but found it as a non-TGEV [3, 7] and the coronavirus-like agent causing the diseases was identified as CV777. The same year, a corona-like virus was reported to be affecting pigs in Belgium [8] while a similar case of corona-like virus infection affecting pig herds was reported in Quebec, Canada in 1980 [9]. Between 1971 and 1980, the virus reportedly spread rapidly in Europe and it was in 1982 when the first outbreak in Asia occurred that caused severe cases of diarrhea in neonate piglets that contributed to high mortality [3]. Several years later, the virus was reported in China [10,11,12,13,14,15,16,17], Japan [18, 19], South Korea [20,21,22], Thailand [23, 24], Vietnam [25, 26] and the Philippines [5, 27] (May 17, 2013, the United States Department). In May 17, 2013, the United States Department of Agriculture (https://www.aphis.USDA2013.gov/animal_health/animal_dis_spec/swine/downloads/ped_tech_note.pdf) National Veterinary Service Laboratory (NVSL) confirmed the first PEDV diagnosis in Iowa, United States [2,3,4, 28, 29] (https://www.aphis.USDA2013.gov/animal_health/animal_dis_spec/swine/downloads/ped_tech_note.pdf). In the same year, the University of Minnesota, Veterinary Diagnostic Laboratory (St. Paul, MN) came up with a report on the detection of a PEDV in a sample from Mexico [30]. Several outbreaks of PED were reported in Canada, Taiwan [31] and Japan in 2013 [3, 18, 19] (https://www.aphis.USDA2013.gov/animal_health/animal_dis_spec/swine/downloads/ped_tech_note.pdf) while reports on the emergence of PED continuously circulates in Europe, Belgium [32], Germany, France, Austria [33] and Slovenia [4].
The disease usually presents with scouring or diarrhea among piglets in the suckling and growing stages. It is characterized by the excretion of watery diarrhea with fetid odor and emaciation in affected pigs. Clinical signs include anorexia, vomiting, diarrhea and severe dehydration [34] which are almost similar to the clinical signs reported for TGEV (https://www.aphis.USDA2013.gov/animal_health/animal_dis_spec/swine/downloads/ped_tech_note.pdf) that can only be differentiated and confirmed through laboratory tests [35]. It is important to note that the disease affects pigs of all ages with morbidity and mortality rate that could be as high as 100% [34] (https://www.aphis.USDA2013.gov/animal_health/animal_dis_spec/swine/downloads/ped_tech_note.pdf). The PEDV spreads slower than TGEV [36]. Clinical signs may occur within four to 5 days following introduction of an infected swine in a herd with susceptible animals. The disease can spread through direct transmission of the PEDV via feco-oral route (https://www.aphis.USDA2013.gov/animal_health/animal_dis_spec/swine/downloads/ped_tech_note.pdf). Indirect transmission is possible through contaminated personnel, equipment and other fomites that enter the herd [36]. Indeed, the PEDV had spread rapidly [3] and caused local epidemics and pandemics. The PED has emerged as one of the most devastating disease of swine, leading to significant losses in the industry [3]. The economic loss involved in PED is directly due to death and production loss, vaccination and compensations in maintaining biosecurity in farms (https://www.aphis.USDA2013.gov/animal_health/animal_dis_spec/swine/downloads/ped_tech_note.pdf).
The PEDV is an enveloped RNA virus that belongs to the genus Alphacoronavirus of the family Coronaviridae [3, 16, 34, 37]. The viral genome is single-stranded, positive-sense RNA. It has six (6) protein genes which include replicase (Rep), spike (S), open reading frame (ORF), envelope (E), membrane (M) and nucleocapsid (N) genes. Like most of the coronavirus, the PEDV has large (20 nm long) club-shaped peplomers and nucleocapsid of helical symmetry [38]. The PEDV is comprised of three (3) major viral proteins: which vary in molecular sizes; the S protein (180–220 kDa), M protein (27–32 kDa) and N protein (55–58 kDa). The S protein plays an important role in viral entry [3, 6, 39] through the viral-cellular fusion activity and induces immune response in the natural host during replication [3, 40]. The M protein is important in the process of virus-assembly [3, 6] and in the induction of protective antibodies with neutralizing activity [3]. The N protein binds viral RNA, provide structural scaffoldings for viral transcription, replication and assembly [6, 41]. The N protein reportedly protects the viral genome during the coronavirus assembly [41]. It also perturbs other antiviral responses by antagonizing interferon production, establishing immune evasion strategy and activation of the NF-κB [3].
In the Philippines, several outbreaks have been recorded in 2005–2006 [42], 2010 and 2014–2015 [5]. The estimated loss during the 2005–2006 PED outbreaks was USD 14.76 million due to piglet mortality and other production-related setbacks [42]. In an epidemiological surveillance on PED conducted by the World Animal Health Organization (OIE) during outbreaks in Asia, the Philippines was reported with the highest number of deaths with approximately 60,000 piglets in 2006 and 2179 piglets in 2007 while there were 39,128 recorded cases of PED in China in 2005 with 2317 deaths (5.92% mortality rate) [42]. In a Philippine-based PED outbreak in 2010, 67% mortality (17,115 heads) of the affected pigs in the province of Batangas was reported. In a surveillance study, it was found that the overall prevalence of PED in the Philippines was higher in small-hold or backyard farms than in the commercial farms [5].
Recognition of the N protein reportedly indicates PED viral replication and its presence is necessary for the early and accurate detection of the virus when the highest expression level of the gene occurs during the early phase of infection [14, 43]. The N gene of PEDV has an approximate size of 1326 bp and has been used by researchers in the diagnosis [44] and in sequence analysis to elucidate the evolutionary relationship of local PEDV strains from those that were reported in the GenBank as this gene is highly conserved [13, 43].Thus, a research work that involved molecular detection of PED virus through RT-PCR and sequence analysis were undertaken to verify the existence of PEDV during the reported outbreak and to reflect on its relationship with other PEDV strains reported in other countries. This can be used as a basis for other PED virus studies and for stakeholders to plan management approach for PED in the country.
Materials and methods
Sample collection
Fecal samples were collected from 23 piglets 1–14 day-old during a PED outbreak in farms located in Bulacan, Nueva Ecija and Tarlac provinces, Central Luzon, Philippines. The farms were chosen based on the reports of high incidence of mortality that ranged from 50 to 100% during the outbreak and the implementation of biosecurity programs allowed the collection of samples once a week for two consecutive weeks. Alive morbid piglets with previous history of a long-standing diarrhea and abdominal distension were chosen as donors of fecal samples while sections of small intestine with extreme thinning of mucosal linings observed during necropsy of piglets were collected for analysis. The samples were stored frozen until use. The PED virus positive control was extracted from the PED oral vaccine (PEDV strain DR13, Green Cross Veterinary Products, Republic of Korea.
Viral RNA extraction and detection through RT-PCR
The viral RNA was extracted from the fecal and macerated intestine samples applying a conventional RNA extraction method that used Qiazol (Qiagen, Germany). The complementary DNA was synthesized using PrimeScript™ II 1st strand cDNA Synthesis Kit (TaKaRa, Japan) following the instruction of the manufacturer.
A pair of primers (Forward, 5′-TGCGGTTCTCACAGATAGTG-3′ and Reverse, 5′-AAGTCGCTAGAAAAACACTCAGTA-3′) targeting the N gene [14] was used in the detection of PED virus. The amplified product recognized by the afore-mentioned primers is 1380 base pairs [14].
The PCR mixture consists of 1.5 µL of the cDNA, 0.05 µL Taq Polymerase (Promega, USA), 0.6 µL 25 mM MgCl2, 0.6 µL 2.5 mM each dNTPs, 0.3 µL Forward primer (10 pmol), 0.3 µL Reverse primer (10 pmol), 2 µL 5 × PCR Buffer and sterile water up a total volume of 10 µL. The thermocycler was programmed to perform at 94 °C for 2 min, followed by 35 cycles of 94 °C for 15 s, 55 °C for 30 s, 72 °C for 45 s, and 72 °C for 7 min with a final hold of 4 °C. The products were resolved in 2% agarose gel after amplification. The PCR products were also sent for sequencing at the Philippine Genomic Center (PGC).
PED virus N gene analysis
The genetic sequence of the N gene was aligned using MEGA 6 software and compared with the sequences of other N gene of PED in the GenBank using the Basic Local Alignment Search Tool (BLAST) program of the National Center for Biotechnology Information (NCBI). A multiple alignment tool (MUSCLE, M6: Alignment Explorer) was used as a reference in determining the phylogenetic relationships of PED strains. A phylogenetic tree was created using the MEGA 6 software where bootstrap values were estimated for 1000 replicates.
Results and discussion
N gene sequence alignment and phylogenetic tree analyses
The involvement of the PED virus in a reported outbreak of the disease in commercial farms located in the provinces of Bulacan, Nueva Ecija and Tarlac, Philippines was explored. Molecular technique such as RT-PCR was applied in the detection and analysis of the partial sequence of the PEDV N gene.
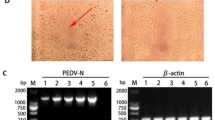
Out of the 34 suspected PEDV infected samples from fecal and intestinal origin, only 10 intestinal samples were detected positive for PED virus (Fig. 1) as demonstrated by the amplification of the 1380 bp segment of the N gene. Field isolates (K3 and K4: Accession No. LC384327, LC384328, respectively) were sequenced and analyzed and data demonstrated that the PED viruses between field samples were 98–99% homologous to PEDV isolates from USA, China, Mexico, Canada and Japan.
Interestingly, the phylogenetic tree analysis revealed that the Philippine samples clustered in group 2–1 of the PEDV composed of the PED strains isolated in USA, Japan, Mexico and Korea (Fig. 2).
Nucleotide and amino acid sequences analyses
Analysis of the partial nucleotide and amino acid sequences revealed mutations, deletions and insertions in the N-gene of the PEDV. Multiple sequence alignment of PEDV samples K3 and K4 showed nucleotide changes at different loci in the N gene relative to the US, China, South Korea, Japan and Vietnam strains. For PED virus sample K3, the following changes were observed: C → G at position 188; G → A at position 192; C → A at position 455; T → A at position 457; A → T at positions 487, 494 and 625; A → G at positions 588, 591 and 616; and A → C at position 610. Deletion was also noted at positions 36, 619, 627 and 708 while insertion was observed at positions 561 and 670 (supplementary Table 1). The PED virus sample K4 on the other hand was observed to have changes in the nucleotides from T → G at position 190 and T → A at position 457. Deletion and insertion were noted at nucleotides 192 and 515, respectively (supplementary Table 2).
The nucleotide changes, deletions and insertions have also lead to changes in the amino acid sequence. Data demonstrate that the N gene of the PEDV K3 had mutation at nine specific positions: P → R at position 188; W → stop codon at position 192; S → stop codon at position 455; W → R at position 457; K → stop codon at position 487; Q → L at positions 494–495; I → M at position 588; position 610 S → R; at position 610; position 616 R → G; and at position 616. Deletion of amino acids in four positions was also detected at 36, 619, 627 and 708 while insertion is observed at position 561 and 670 (supplementary Table 3). Slight changes in the amino acid were observed in PEDV K4. Data demonstrate that PEDV K4 had mutation from W → R at position 457, deletion at position 190–192 and an insertion at position 515 (supplementary Table 4).
The swine industry plays a major role in harnessing food security for the Filipinos. The industry includes more than 12 million heads which accounts for an 80% of the total livestock production each year [45]. Animal stocks are threatened with many diseases that vary from bacterial to viral in etiologies like the PEDV [5, 42]. The PED is considered as one of the most devastating diseases affecting the swine industry in the Philippines. Management and control schemes including biosecurity measures and vaccination have been reportedly applied. However, disappearance and re-appearance of PED can be an indication of failures in vaccination, biosecurity and control systems [34]. Sporadic cases or outbreaks of PED reportedly occur in different locations of the country and one possible reason for PED emergence is the introduction of new PEDV strains or even multiple strains through movement of stocks from one country to another [34].
According to researches, the PEDV is clustered into two groups: the classical PEDV strains that first emerged in 1970s in Europe and the highly virulent PEDV strains that emerged in 2010 in China [3, 17]. The G1 PEDV strains are the classical PEDV, which can be clustered further into G1a where the prototype vaccine strain CV777 belongs; and G1b which includes new PED variants identified in South Korea, China, US and Europe [3]. The G1b are believed to be a result of recombination of the classical G1a and G2 PEDV strains. The G2 PEDV strains or the “emerging” strains purportedly form clusters which include the S-indel and non-S-indel. The G2 PEDV strains are the virulent strains that cause local epidemics in Asia and pandemics in the US and Europe [3]. The G2 PEDV strains are described with notable genetic sequence with insertions and deletions in the S gene [3]. The S-indel cluster is reportedly composed of the PEDV strains that caused outbreaks in China, Japan, Korea, US and Europe and this group are claimed to be less virulent [3, 30, 37, 46]. The G2b cluster or the non-S-indel include the virulent PEDV strains that were isolated in China and North America which were believed to be the descendants of China PEDV strains that underwent processes of recombination [3, 30, 37, 46].
Though some researchers reported inconsistencies in the phylogenetic tree analysis using specific genes as compared to the whole genome sequence [16], these findings are still significant in designing control measures, biosafety, vaccine production and development of diagnostic tools as selection of the target gene depends mainly on the purpose of the study [3, 16]. The researchers suggest that in determining the relatedness of PEDV strains, the S gene is inappropriate since the phylogenetic tree structure is similar or closest to the structure that uses the whole genome sequence. The M and N genes are reportedly conserved in PEDV. Some researchers however suggested that the N gene is a choice in the development of diagnostic protocols such as RT-PCR in detecting various strains of PEDV since this is a more conserved gene [13, 43]. It is also important to note that the N gene can differentiate PEDV from other closely related viruses such as the TGEV [47] and can determine whether a PEDV strain is an exotic or a product of continuous evolution pressures in the original variant [3].
Interestingly, the phylogenetic analysis of the N gene based on previous studies [29] pointed out that the two Philippine PEDV samples belong to PEDV G2 which is in concordance with another report that used sequence analysis of ORF3 genes [27]. Two (2) substitutions in the SS6 epitope domain of the Philippine PED strains and the SS6 domain region of PED strains from other countries and one to three amino acid mutations were also reported in the analysis of the ORF3 gene [27]. However, the partial sequence analysis of the S gene in samples from the Philippines revealed that the PEDV which predominates in the Philippines are clustered into two groups [5]. This result can address the questions of swine practitioners on the constant re-emergence of in the Philippines in spite of vaccination and other control programs. This triggers moderate concern for the reason that a PED strain with a known genetic background when used in the preparation of a vaccine may not offer protection to the herd afflicted with PED with a different strain. Currently, the only PEDV vaccine in the Philippines is the DR13 oral vaccine, which was derived from a different group based on phylogenetic tree analysis. The efficiency of a vaccine in protecting a herd is influenced by its antigenic, genetic (> 10% amino acid variation between respective S proteins) and phylogenetic (G1 vs. G2) properties not different from the field epidemic strains [20]. This then implies that PEDV strains prevalent in the field or in a locality have to be used for the development of next generation vaccines to control PED [3].
Currently, no country has reported eradication of PED and appropriate measures have been recommended to control the spread of the disease. It is noteworthy that PED is not an OIE-listed disease, considered not-reportable and no quarantine or movement restrictions for PED is described internationally (https://www.aphis.USDA2013.gov/animal_health/animal_dis_spec/swine/downloads/ped_tech_note.pdf). However, control of PED spread is not only limited to implementation of certain biosecurity measures but also through the capability of a country to detect or diagnose PED properly [42]. Equally important is the gathering of relevant information related to the epidemiology and the capability of the country to detect and diagnose the disease to trace-back infections [3, 5, 6, 18, 24, 42]. Clear understanding of the origin and transmission of the PEDV may contribute to a more effective prevention and control of the disease including biosecurity [3, 25, 34, 42]. It is recommended that further studies and analysis be conducted to fully elucidate the PEDV strain or strains in the Philippines. A comprehensive molecular and pathological evaluation are equally important in the development of vaccine and diagnostic protocols for PEDV [3, 26, 28, 29, 34].
References
Song DS, Oh JS, Kang BK, Yang JS, Song JY, Moon HJ, Kim TY, Yoo HS, Jang YS, Park BK. Fecal shedding of a highly cell-culture-adapted porcine epidemic virus after oral inoculation in pigs. J Swine Health Prod. 2005;13(5):269–72.
Wang S, Cheng X, Chen S, Lin F, Jiang B, Zhu X, Li Z, Wang J, Chen S. Classification of emergent US strain of PED virus by phylogenetic analysis of nucleocapsid and ORF3 genes. J Clin Microbiol. 2014;52(9):3509–10.
Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J. 2015;12:93.
Toplak I, Ipavec M, Kuhar U, Kusar D, Papic B, Koren S, Toplak N. Complete genome sequence of the porcine epidemic diarrhea strain SLO/JH-11/2015. Genome Announc. 2016;4(2):e01725-15.
Paraguison-Alili R, Domigo CYJ. Phylogenetic tracking of current porcine epidemic diarrhea virus (PEDV) strains in the Philippines. Arch Virol. 2016;161(9):2601–4.
Teenavechyan S, Frantz PN, Wongthida P, Chailangkarn T, Jaru-ampornpan P, Koonpaew S, Jongkaewwattana A. Deciphering the biology of porcine epidemic diarrhea virus in the era of reverse genetics. Virus Res. 2016;226:152–71.
Wood EN. An apparently new syndrome of porcine epidemic diarrhea. Vet Rec. 1977;100:243–4.
Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978;50:243–7.
Turgeon DC, Morin M, Jolette J, Higgins R, Marsolais G, Di Franco E. Coronavirus-like particles associated with diarrhea in baby pigs in Quebec. Can Vet J. 1980;21:100–23.
Fan H, Zhang J, Ye Y, Tong T, Xie K, Lao M. Complete genome sequence of novel porcine epidemic diarrhea virus in South China. J Virol. 2011;86(18):10248–9.
Chen F, Pan Y, Zhang X, Tian X, Wang D, Zhou Q, Song Y, Bi Y. Complete genome sequence of a variant porcine epidemic diarrhea virus strain isolated in China. J Virol. 2012;86(22):12448.
Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis. 2012;18(1):161–3.
Chen J, Liu X, Shi D, Shi H, Zhang X, Li C, Chi Y, Feng L. Detection and molecular diversity of Spike gene of porcine epidemic diarrhea virus in China. Viruses. 2013;5:2601–13.
Li Z, Chen F, Yuan Y, Zeng X, Wei Z, Zhu L, Sun B, Xie Q, Cao Y, Xue C, Ma J, Bee Y. Sequence and phylogenetic analysis of nucleocapsid genes of porcine epidemic diarrhea virus (PEDV) strains in China. Arch Virol. 2013;158:1267–73.
Chen F, Zhu Y, Wu M, Ku X, Ye S, Li Z, Guo X, He Q. Comparative genomic analysis of classical and variant virulent parental/attenuated strains of porcine epidemic diarrhea virus. Viruses. 2015;7:5525–38.
Zhang J, Chen Q, Gauger JQ, Harmon KM, Yoon KJ. Reply to “Classification of Emergent US strain of PED virus by phylogenetic analysis of nucleocapsid and ORF3 genes”. J Clin Microbiol. 2014;52(9):3511–4.
Li R, Tian X, Liu Y, Xu J, Liu D. Isolation and genetic analysis of a variant porcine epidemic diarrhea virus in China. Pol J Vet Sci. 2016;19(1):65–73.
Murakami S, Miyazaki A, Takahashi O, Hashizume W, Hase Y, Ohashi S, Suzuki T. Complete genome sequence of the porcine epidemic diarrhea virus variant Tottori2/JPN/2014. Genome Announc. 2015;3(4):e00877-15.
Diep NV, Norimine J, Sueyoshi M, Lan NT, Hirai T, Yamaguchi R. US-like isolates of epidemic diarrhea virus from Japanese outbreaks between 2013 and 2014. Springerplus. 2015;4:756.
Lee S, Lee C. Outbreak-realted porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg Infect Dis. 2013;20(7):1223–6.
Cho YY, Lim SI, Kim YK, Song JY, Lee JB, An DJ. Complete genome sequence of K14J1301, a novel variant strain of porcine epidemic diarrhea virus in South Korea. Genome Announc. 2014;2(3):e00505-14.
Lee S, Park GS, Shin JH, Lee C. Full-genome sequence analysis of variant strain of porcine epidemic diarrhea virus in South Korea. Genome Announc. 2014;2(6):e01116-14.
Cheun-Arom T, Temeeyasen G, Srijangwad A, Tripipat T, Sangmalee S, Vui T, Chusana T, Tantituvanoat A, Nilubol D. Complete genome sequence of two genetically distinct variants of porcine epidemic diarrhea virus in the Eastern Region of Thailand. Genome Announc. 2015;3(3):e00634-15.
Stott CJ, Wiratsudakul A, Temeeyasen G, Sawattrakool K, Nilubol D. An introduction of porcine epidemic diarrhea virus to Thailand. Thai J Vet Med Suppl. 2016;46:369–70.
Duy DT, Toan NT, Puranaveja S, Thanawongnuwech R. Genetic characterization of porcine epidemic diarrhea virus (PEDV) isolates from Southern Vietnam during 2009–2010 outbreaks. Thai J Vet Med. 2011;41(1):55–64.
Vui DT, Tung N, Inui K, Slater S, Nilubol D. Complete genome sequence of porcine epidemic diarrhea virus in Vietnam. Genome Announc. 2014;2(4):e00753-14.
Kim YY, Cho YY, An BH, Lim SI, Lim JA, Cho IS, Le VP, An DJ. Molecular characterization of the spike and ORF3 genes of porcine epidemic diarrhea virus in the Philippines. Arch Virol. 2016;161:1323–8.
Marthaler D, Bruner L, Collins J, Rossow K. Third strain of porcine epidemic diarrhea virus, United States. Emerg Infect Dis. 2014;20(12):2162–3.
Wang L, Byrum B, Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg Infect Dis. 2014;20(5):917–9.
Vlasova AN, Marthaler D, Wang Q, Culhane MR, Rossow KD, Rovira A, Collins J, Saif LJ. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg Infect Dis. 2014;20(10):1620–8.
Lin CN, Chung NB, Chang SW, Wen CC, Liu H, Chien CH, Chiou MT. US-like strain of porcine epidemic diarrhea virus outbreaks in Taiwan, 2013–2014. J Vet Med Sci. 2014;76(9):1297–9.
Theuns S, Conceição-Neto N, Christiaens I, Zeller M, Desmarets LM, Roukaerts ID, Acar DD, HeylenE, Matthijnssens J, Nauwynck HJ. Complete genome sequence of a porcine epidemic diarrhea virus from a novel outbreak in Belgium. Genome Announc. 2015;3(3):pii:e00506-15.
Steinrigl A, Fernandez SR, Stoiber F, Pikalo J, Sattler T, Schmoll F. First detection, clinical presentation and phylogenetic characterization of porcine epidemic diarrhea virus in Austria. BMC Vet Med. 2015;11:310.
Jung K, Saif LJ. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J. 2015;204(2):134–43.
Song D, Park B. Porcine epidemic diarrhea virus: a comprehensive review of molecular epidemiology, diagnosis and vaccines. Virus Genes. 2012;44:167–75.
Pospischil A, Stuedli A, Kiupel M. Diagnostic notes: update on porcine epidemic diarrhea. J Swine Health Prod. 2002;10:81–5.
Lin CM, Saif LJ, Marthaler D, Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39.
Murphy F, Gibbs E, Horzinek M, Studdert M. Veterinary virology. 3rd ed. London: Academic Press; 1999.
Bosch BJ, Vann der Zee R, de Han CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of fusion core complex. J Virol. 2003;77(16):8801–11.
Klumperman J, Locker JK, Meijer A, Horzinek M, Geuze HJ, Rottier PJ. Corona virus M proteins accumulate in the golgi complex beyond the site of virion budding. J Virol. 1994;68(10):6523–34.
Spaan W, Delas H, Skinner M, Arstrong J, Rottier P, Smeekens S, van der Zeist BA, Siddel SG. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2(10):1839–44.
Morales RG, Umandal AC, Lantican CA. Emerging and re-emerging disease in Asia and the Pacific with special emphasis on porcine epidemic diarrhea. Conf OIE. 2008;2007:185–9.
Sun P, Gu CY, Ding YY, Wei JH, Yin ZJ, Geng ZJ, Li Y. Sequence and phylogenetic analyses of the M and N of porcine epidemic diarrhea virus (PEDV) strains in Anhui Province, China. Genet Mol Res. 2015;14(4):13403–13.
Kubota S, Sasaki O, Amimoto K, Okada N, Kitazima T, Yasuhara H. Detection of porcine epidemic diarrhea virus using polymerase chain reaction and comparison of the nucleocapsid protein genes among strains of the virus. J Vet Med Sci. 1999;61(7):827–30.
BAS. Swine performance report, January to June 2013. Bureau of Animal Statistics, Philippines. 2014; http://www.AgriWorld.nl. Accessed 02 Aug 2014.
Liu XS, Wang Q. Reverse transcription PCR assays for the differentiation of various US porcine epidemic diarrhea virus strains. J Virol Methods. 2016;234:137–41.
Diel DG, Lawson S, Okda F, Singrey A, Clement T, Fernandes MHV, Christopher-Hennings J, Nelson EA. Porcine epidemic diarrhea virus: an overview of current virological and serological diagnostic methods. Virus Res. 2016;226:60–70.
Acknowledgements
The authors would like to thank Dr. Arnel N. Del Barrio, Executive Director of the Philippine Carabao Center (PCC), for the support to complete the study. Special thanks to all the staff of the Biosafety and Environment Section of PCC for their technical support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Garcia, G.G., Aquino, M.A.D., Balbin, M.M. et al. Characterisation of porcine epidemic diarrhea virus isolates during the 2014–2015 outbreak in the Philippines. VirusDis. 29, 342–348 (2018). https://doi.org/10.1007/s13337-018-0470-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-018-0470-4