Abstract
Limited therapeutic efficacy of temozolomide (TMZ) against glioblastomas highlights the importance of exploring new drugs for clinical therapy. Sunitinib, a multitargeted receptor tyrosine kinase inhibitor, is currently being tested as therapy for glioblastomas. Unfortunately, sunitinib still has insufficient activity to cure glioblastomas. Our aim was to determine the molecular mechanisms counteracting sunitinib drug sensitivity and find potential adjuvant drugs for glioblastoma therapy. Through in vitro experiments, transcriptome screening by RNA sequencing, and in silico analyses, we found that sunitinib induced glioma apoptotic death, and downregulated genes were enriched in oncogenic genes of glioblastoma. Meanwhile, sunitinib-upregulated genes were highly associated with the protective autophagy process. Blockade of autophagy significantly enhanced sunitinib’s cytotoxicity. Growth arrest and DNA damage-inducible protein (GADD) 34 was identified as a candidate involved in sunitinib-promoted autophagy through activating p38-mitogen-activated protein kinase (MAPK) signaling. Higher GADD34 levels predicted poor survival of glioblastoma patients and induced autophagy formation in desensitizing sunitinib cytotoxicity. Guanabenz, an alpha2-selective adrenergic agonist and GADD34 functional inhibitor, was identified to enhance the efficacy of sunitinib by targeting GADD34-induced protective autophagy in glioblastoma cells, TMZ-resistant cells, hypoxic cultured cells, sphere-forming cells, and colony formation abilities. A better combined treatment effect with sunitinib and guanabenz was also observed by using xenograft mice. Taken together, the sunitinib therapy combined with guanabenz in the inhibition of GADD34-enhanced protective autophagy may provide a new therapeutic strategy for glioblastoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma (GBM), a grade IV glioma, is divided into 3 distinct types, including IDH–wild type (about 90% patients) which usually occurred at elder age (~ 62 years old), and most of them belong to de novo or primary GBM; IDH–mutant (closely to secondary GBM) with 10% patients in a younger age (~ 44 years old) through malignant progression from lower-grade diffuse glioma; and not otherwise specified (NOS) with no IDH type evaluation performed [1, 2]. Currently, the major therapy for GBM is surgery followed by radiation and temozolomide (TMZ) chemotherapy. However, the GBM patients after treatment only have a median overall survival of around 15 or 31 months for IDH1–wild type or mutant type respectively. And an effective supportive care for monitoring and managing the neurological complications such as cerebral edema or seizures was also included. Due to the rapid proliferative and highly invasive nature of glioma cells, these therapies have limited therapeutic efficacy [3, 4]. During the malignant transformation of GBM, abnormally expressed growth factors, such as platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF), promote glioma cell proliferation, invasion, and angiogenesis by interacting with their receptors [5, 6]. Therefore, a variety of receptor tyrosine kinase (RTK) inhibitors were designed to target these growth factors and their receptors [7]. Although these drugs exhibited initial success in preclinical experiments, they only show limited efficacy in clinical trials [8,9,10]. Hence, it is important to clarify the underlying mechanisms that impede the therapeutic efficacy of RTK inhibitors in GBM.
Sunitinib, a multitargeted receptor tyrosine kinase inhibitor (TKI), suppresses KIT, the PDGF receptor (PDGFR), and VEGF receptor (VEGFR). Sunitinib acts as a toxin towards proliferating endothelial cells and tumor vessels in gliomas [11]. Sunitinib also induces glioma cell death and suppresses invasion in vitro [12, 13]. Furthermore, sunitinib exhibits an antiangiogenic effect and prolongs the survival time in mice bearing intracerebral GBM tumors [13]. Sunitinib sensitizes the GBM cells to TMZ treatment [14], and significantly improves its response in O6-methylguanine-DNA methyl-transferase (MGMT)-positive cells [15]. Martinho et al. [16] also report that inhibition of AXL, a receptor tyrosine kinase (RTK), in GBM cells is less sensitive to sunitinib cytotoxicity. Despite the promising effects of sunitinib on suppressing glioma growth, sunitinib therapy still fails to achieve clinical improvement in GBM patients. Single-agent treatment of sunitinib at 37.5 mg/day with progressive high-grade glioma patients had insufficient activity in reducing tumor growth [17]. The median overall survival was 3.8 (95% CI 2.2-5.3) months. After examining the clinical outcomes of glioma patients, there was no association between sunitinib target protein expressions and its efficacy. Similarly, no single-agent objective antitumor activity of sunitinib was observed in child patients with recurrent or refractory high-grade glioma [18] and patients with nonresectable [10] or recurrent GBM [19]. In addition, a phase I study of sunitinib and irinotecan (a topoisomerase-1 inhibitor) combination with recurrent GBM patients also occupied moderate toxicity and limited antitumor activity [20]. Hence, there are other unidentified mechanisms that might be involved in the poor efficacy of sunitinib. Previous studies indicated that induction of autophagy is involved in affecting sunitinib efficacy in different kinds of cancers such as renal cell carcinoma [21], prostate cancer [22], and ovarian cancer [23]. However, whether autophagy is associated with the poor efficacy of sunitinib in gliomas remains unclear.
Autophagy is activated by different kinds of stressors to promote the production of nutrients and disassemble dysfunctional organelles [24]. Therefore, the function of autophagy is to maintain cancer cell survival under harsh microenvironments such as hypoxia and nutrient depletion [25]. Besides tumor microenvironment-derived cues, chemotherapeutic drugs and molecularly targeted therapies also create stress conditions that can trigger autophagy induction by oxidative phosphorylation or by inhibiting vascularization [26, 27]. As for gliomas, TMZ and bevacizumab promote protective autophagy through inhibiting Akt-mammalian target of rapamycin (mTOR) signaling [28, 29]. Further, the cytotoxic effects of both drugs are augmented after combining with the autophagy inhibitor, chloroquine (CQ) [30, 31]. Therefore, the autophagic process plays an important role in the formation of drug resistance in gliomas. Sunitinib, which is a lysosomotropic drug, can diffuse into lysosomal membranes and is protonated because of low pH values. Thus, sunitinib is trapped in lysosomes because of the loss of the ability to diffuse through the membrane. This further leads to autophagic induction in pancreatic neuroendocrine tumors [32]. Nevertheless, few studies have reported possible mechanisms involved in sunitinib-induced autophagy in gliomas. Understanding the roles and underlying mechanisms that lead to autophagic formation in gliomas could further guide us in developing new strategies to improve sunitinib’s therapeutic efficacy.
The growth arrest and DNA damage-inducible protein (GADD34) is a regulatory subunit of protein phosphatase 1, and is upregulated under different stress conditions such as amino acid deprivation, DNA damage, and endoplasmic reticular (ER) stress [33]. One study reported that GADD34 promotes autophagy in liver cells under starvation [34]. The mechanism is through its interaction with tuberous sclerosis complex 2 (TSC2) by dephosphorylation of phosphorylated (p)-TSC2 at Thr1462 [34]. The dephosphorylation of p-TSC2 inhibits mTOR singling, leading to autophagy formation [34]. Another study demonstrated that GADD34 depletion or use of the GADD34 inhibitor, guanabenz, an alpha2-selective adrenergic agonist used as an antihypertensive agent, suppresses autophagy-mediated survival of liver cancer cells [35]. These findings suggest that GADD34 participates in promoting a protective autophagic process. However, the role of GADD34 in gliomas remains poorly understood.
In the present study, we aimed to determine the molecular mechanisms that counteract sunitinib drug sensitivity to GBM cells. By targeting this key molecular pathway, we evaluated the potential adjuvant drug for combined treatment with sunitinib for GBM therapy. By performing an RNA sequencing (RNA-Seq) analysis, we established a sunitinib-mediated transcriptome profile in GBM. We found that sunitinib-upregulated genes were significantly enriched in the autophagic process. We also identified that sunitinib induced protective autophagy by desensitizing its cytotoxicity. We found that GADD34, a risk gene for predicting poor survival of GBM patients, was upregulated by sunitinib-mediated p38-MAPK signaling, resulting in autophagy induction. Finally, we identified that guanabenz significantly enhanced the efficacy of sunitinib by targeting colony formation and GADD34-induced protective autophagy in GBM cell lines, TMZ-resistant cells, 1% hypoxia-cultured cells, and tumorsphere cells. A better combined treatment effect with sunitinib and guanabenz was also observed using xenograft mice. We concluded that combination therapy with sunitinib and guanabenz, which inhibits GADD34-induced autophagy, may provide a new strategy for future GBM therapy.
Materials and Methods
Chemicals and Reagents
The human glioblastoma U87-MG cell line was purchased from the Bioresource Collection and Research Center (Hsinchu City, Taiwan). T98G, A172, and HCM-BROD-0106-C71 (ATCC® PDM-123™) cells were purchased from American Type Culture Collection (ATCC; Rockville, MD). Reagents for cell culture were purchased from GIBCO-BRL (Grand Island, NY). Anti-GADD34 (cat. no. GTX115747), Ki67 (cat. no. GTX16667), caspase-3 (cat. no. GTX110543), poly(ADP ribose) polymerase (PARP; cat. no. GTX100573), light chain 3 (LC3)-I/LC3-II (cat. no. GTX127375), p62 (cat. no. GTX100685), ATG12/ATG5 (cat. no. GTX124181), beclin-1 (cat. no. GTX133555), p-p38 (cat. no. GTX24822), p38 (cat. no. GTX110720), LAMP1 (cat. no. GT25212), and anti-β-actin (cat. no. GTX109639) antibodies were purchased from GeneTex (Hsinchu City, Taiwan). An FITC Annexin V Apoptosis Detection Kit (cat. no. 556547) was purchased from Becton Dickinson (Franklin Lakes, NJ). An enhanced chemiluminescence (ECL) solution (cat. no. WBKLS0500) and polyvinylidene difluoride (PVDF) membranes (cat. no. IPVH00010) were purchased from Millipore (Billerica, MA). Acridine orange (AO; cat. no. A9231), sunitinib (cat. no. PZ0012), TMZ (cat. no. T2577), CQ diphosphate (CQ; cat. no. C6628), hydroxychloroquine (HCQ; cat. no. H0915), Lys05 (cat. no. 2097), cycloheximide (CHX; cat. no. 01810), and guanabenz (cat. no. G110) were purchased from Sigma-Aldrich (St. Louis, MO). Trizol® reagent (cat. no. 15596026), Lipofectamine 3000 (cat. no. L3000015), and secondary antibodies (cat. no. A16110) were purchased from Invitrogen (Carlsbad, CA). SYBR® Green polymerase chain reaction (PCR) master mix (cat. no. 4309155) and a MultiScribe (tm) reverse-transcriptase kit (cat. no. N8080234) were purchased from Applied Biosystems (Waltham, MA). Scrambled short hairpin (sh)RNA and GADD34 shRNA were purchased from the National RNAi Core Facility (Nankang, Taipei, Taiwan). Unless otherwise specified, all other reagents were of analytical grade.
Cell Culture, Generation of TMZ-Resistant Cells, Drug Treatment, and Gene Transfection
U87-MG, U87-R, A172, and T98G-R cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 2.5 mM GlutaMAX, 100 units/mL penicillin, 100 μg/mL streptomycin, 1 mM sodium pyruvate, and 10% fetal bovine serum (FBS; Biological Industries, Cromwell, CT). For establishing the TMZ-resistant cells, 104 U-87 MG and T98G parental cells per well were respectively seeded into 96-well plates. In the first step, a low dose of TMZ (10 μM) was used to maintain the cells. After the surviving cells grew stably for 15 days, the dose of TMZ was gradually increased (10 to 600 μM). Finally, the IC50 values of TMZ in these 2 cells were evaluated by MTT assays. The induced TMZ-resistant cells were named as U87-R and T98G-R. Twenty micromoles of TMZ was also added when culturing U87-R and T98G-R cells. The method for culturing HCM-BROD-0106-C71 cells was according to the instruction from ATCC. The Ultra Low Attachment (ULA) flasks/plates (cat. no. 3814; Corning, Corning, NY) were used. The NeuroCult NS-A medium (cat. no. 05750; StemCell Technologies Inc., Vancouver, BC, Canada) with NS-A proliferation supplement (cat. no. 05754; StemCell Technologies), 20 ng/mL EGF (cat. no. 78003.1 StemCell Technologies), 20 ng/mL bFGF (cat. no. 78003; StemCell Technologies), and 2 μg/mL Heparin (cat. no. 07980; StemCell Technologies) was used to maintain HCM-BROD-0106-C71 cell growth. All cells were cultured at 37 °C in a 5% CO2 incubator.
For gene transfection experiments, Lipofectamine 3000 was utilized based on the manufacturer’s protocol. Briefly, cells were seeded in 12- or 6-well plates at a density of around 70%. After overnight culture, cells were respectively transfected with the indicated dose of pCDH-GADD34, empty pCDH, GADD34 shRNA, and scrambled shRNA (cat. no. CD510B-1; System Biosciences, Palo Alto, CA). The pCDH-GADD34 construct was established in our lab.
For drug treatment, indicated concentrations of TMZ, sunitinib, and guanabenz were used overnight or for 72 h. For lysosome inhibitor treatment, the cells were treated with the indicated dose of sunitinib or guanabenz for 24 h after pretreating with lysosome inhibitors for 1 h. For drug treatment in hypoxic conditions, the cells were cultured in an Eppendorf® galaxy® 48R CO2 chamber (Hamburg, Germany) with 1% O2 and 5% CO2.
Cell Viability Assay
A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to measure cell viability. Cells were seeded on a 96-well plate (104 cells/well), followed by drug treatment or gene transfection. Before the end of treatment, 0.5 mg/mL MTT was added to each well for 4 h. After the supernatants had been carefully aspirated out, dimethyl sulfoxide (DMSO) was used to dissolve the formazan crystals. The absorbance was measured at 570 nm with a Thermo Varioskan Flash reader (Carlsbad, CA).
Flow Cytometry
Apoptosis and acidic vesicular organelle detection were respectively analyzed using flow cytometry with annexin V/propidium iodide (PI) double staining or AO. In brief, to detect apoptosis, whole sunitinib-treated cells were collected in buffer containing 10 mM HEPES (pH 7.4), 140 mM NaCl, and 2.5 mM CaCl2. Subsequently, cells were stained with an antibody against annexin V (2.5 μg/mL) and PI (2 ng/mL) for 20 min. The sum of early apoptotic (annexin V+/PI−) and late apoptotic (annexin V+/PI+) cells compared to control groups represented the apoptotic ratio. For acidic vesicular organelle detection, the cells were supplemented with 1 μg/mL AO and incubated in a dark room for 15 min. Then, the percentage of cells with red fluorescence represented the degree of acidic vesicular organelle formation. For each flow cytometric experiment, 104 cells were detected using a flow cytometer with CellQuest software (Becton Dickinson).
Immunoblot Assays
After treatment, total cell lysates were collected and dissolved in ristocetin-induced platelet agglutination buffer containing 0.5% deoxycholate, 1% Nonidet P-40, and 0.1% sodium dodecylsulfate (SDS) in phosphate-buffered saline (PBS) with a protease inhibitor cocktail (cat. no. 539131; Calbiochem, Billerica, MA). After centrifugation at 13,800 × g and 4 °C for 10 min, protein samples were denatured with a buffer containing 2% SDS, 10 mM dithiothreitol, 60 mM Tris-hydrochloric acid (Tris-HCl, pH 6.8), and 0.1% bromophenol blue. Polyacrylamide-SDS gels (10% and 15%) were used to analyze target proteins whose molecular weights were > 50 and < 50 kDa. After transfer onto a PVDF membrane (cat. no. 539131, IPVH00010; Merck, Burlington, MA), PBS containing 5% nonfat dry milk was used for blocking for 1 h at room temperature. Then, the membrane was incubated in PBS containing a primary antibody (diluted 1:1000) overnight at 4 °C. After washing with PBS-Tween 20, the secondary antibody conjugated to horseradish peroxidase (diluted 1:3000) was added to the membrane for 1 h at 25 °C. Antibody–protein complexes were detected with an enhanced chemiluminescence (ECL) nonradioactive detection system.
Next Generation Sequencing
Biological replicates of sunitinib-treated and untreated U-87 MG cells were used for RNA-Seq analyses. Total RNA was extracted using the Trizol® reagent (Invitrogen) according to the manufacturer’s instructions. Cell lysates were sent to Welgene Biotechnology (Taipei, Taiwan) for RNA-Seq services. Read counts were utilized to perform differentially expressed gene (DEG) analyses using edgeR. Gene candidates were considered differentially expressed with |log2 fold change| of ≥ 1 and a false detection rate (FDR) of < 0.01.
Gene Set Enrichment Analysis
A gene set enrichment analysis (GSEA) algorithm with 1000 permutations was conducted using the fgsea package. Genes were ranked based on their multiples of change in different phenotypes. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was selected as the gene set for analyses. Pathways with an FDR of < 0.05 were considered significant.
The Cancer Genome Atlas and Gene Expression Omnibus Data Analyses
Level 3 microarray data of The Cancer Genome Atlas (TCGA) GBM patients (gdac.broadinstitute.org_GBM.mRNA_Preprocess_Median.Level_3) were retrieved and downloaded from the Broad Institute Firehouse (http://gdac.broadinstitute.org/), which contained gene expression profiles and overall survival information of 524 GBM patients. GSE13041 (n = 267) and GSE108474 (Rembrandt; n = 210) microarray data were respectively downloaded from the Gene Expression Omnibus (GEO) database. All of the microarray data were robust multichip average (RMA)-normalized and log2-transformed.
RNA Extraction and Real-Time Reverse-Transcription Quantitative Polymerase Chain Reaction
Total RNA was extracted from treated cells using Trizol® according to the manufacturer’s instructions. RNA quality was checked by A260/A280 readings. Using a MultiScribe (tm) Reverse Transcriptase kit with random primers, complementary (c)DNA was synthesized from 1 μg of total RNA, diluted 1:30 with PCR-grade water, and stored at − 20 °C. Specific primers used for detecting human GADD34 and GAPDH levels were as follows: GADD34-F: ATGATGGCATGTATGGTGAGC, GADD34-R: AACCTTGCAGTGTCCTTATCAG, GAPDH-F: GTGAAGGTCGGAGTCAAC, and GAPDH-R: GTTGAGGTCAATGAAGGG. Gene expression levels were measured using an Applied Biosystems StepOnePlus™ System (Thermo Fisher Scientific, Waltham, MA) with preoptimized conditions. Each PCR was performed in triplicate, and contained 5 μL 2× SYBR Green PCR Master Mix, 0.2 μL primer sets, 1 μL cDNA, and 3.6 μL nucleotide-free H2O to yield a 10-μL reaction. The normalized CT difference was calculated as the expression rate between the control and sample after adjusting for the amplification efficiency relative to the expression level of the GAPDH housekeeping gene.
Tumor Sphere Formation Assay
For tumor sphere formation assays, 3 × 104 cells were cultured in a Costar® ultralow attachment multiple 96-well plate (Corning) with DMEM, B27 (1:50; Invitrogen, San Diego, CA), epidermal growth factor (EGF; 20 ng/mL, Invitrogen), basic fibroblast growth factor (bFGF; 20 ng/mL, Gibco), penicillin/streptomycin (Invitrogen), and l-glutamine (Invitrogen). Spheres were observed on day 4 after plating. One-third of the medium was replaced every 3 days. After 1 week, spheres were dissociated and replated in nonadherent plates to generate secondary spheres. After 2 weeks of culture, the spheres were treated with indicated drugs for further studies.
Soft Agar Colony Formation Assay
Cells (104) were seeded in 12-well plates coated with 2 layers of different agar concentrations. The bottom and top layers respectively consisted of 0.5% and 0.3% agar/DMEM. DMEM (200 μL) with 10% FBS and treated drugs was added to each well to avoid evaporation from the top layer. After culturing for 14 days, colonies were stained and counted with 1 mg/1 mL nitrotetrazolium blue chloride (Sigma-Aldrich).
In Vivo Xenograft Study
All animal care and experimental procedures approved by the Taipei Medical University Laboratory Animal Care and Use Committee (Permit Number: LAC-2019-0115) complied with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH Publications No. 8023, revised 1978) and ARRIVE guidelines. Six-week-old male nude mice (NOD.CB17-Prkdcscid/NcrCrl; BioLASCO Taiwan, Nankang, Taipei, Taiwan) were allowed free access to food and water in a specific pathogen-free room with a light/dark cycle of 12/12 h. U87 MG cells (5 × 107) in 0.1 mL minimum essential medium (MEM) containing diluted 2:1 Corning® Matrigel® Basement Membrane Matrix (cat. no. 354248, Corning) were subcutaneously (s.c.) injected into the right hind flank of mice. Tumor sizes were measured and calculated as 1/2 × length × width2 (mm3) using calipers. When the tumors had grown to about 100 mm3, tumor-bearing mice were randomly separated into 4 groups (n = 5 mice/group), containing the vehicle control (20% DMSO), sunitinib (50 mg/kg), guanabenz (10 mg/kg), and combined treatment groups. The doses of sunitinib and guanabenz used in the present study were according to previous studies [36, 37]. Drugs were peritumorally injected 3 times on days 14, 21, and 28. Tumor sizes and body weights were respectively measured every 3 days. At the end of the experiment, all mice were sacrificed. Tumor tissues were excised for immunohistochemical (IHC) staining and homogenized to extract proteins for further investigation.
Immunohistochemistry
For IHC staining, 5-μm paraffin-embedded sections mounted on poly-l-lysine-coated slides were baked at 60 °C overnight. Then, sections were heated in a microwave oven for 10 min with 10 mmol/L sodium citrate buffer (pH 6) after dewaxing and hydration. After blocking with 10% horse serum in PBS for 30 min, sections were incubated overnight at 4 °C with an anti-Ki67 primary antibody at a dilution of 1:100. Next, biotinylated goat anti-rabbit immunoglobulin G (IgG) at a dilution of 1:1000 was applied for 30 min, followed by detection of immunoreactivity with an avidin–biotin system using 3,3′-diaminobenzidine tetrahydrochloride as a chromogen. Sections were lightly counterstained with Mayer’s hematoxylin (Richard-Allan Scientific, Kalamazoo, MI). Negative controls without primary antibodies were performed for all reactions (data not shown).
Fluorescent Immunocytochemistry
The 104 U-87 MG cells were cultured in gelatin-coated sterilized coverslips. After drug treatment and removing the culture mediums, the cells were washed twice with PBS. Then, the cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. After washing, the cells were blocked with 1% BSA and 0.1% Triton® X-100 for 1 h. The primary antibody (1:100) and secondary antibody with fluorescent conjugate (1:300) in 1% BSA buffer were used to incubate the cells at room temperature for 1 h. After repeated washing, the mounting buffers containing DAPI were used. Finally, coverslips were placed on the microscope slide. The flourescent immunocytochemistry (FICC) data were captured by using a confocal microscope.
Autophagy Detection
According to autophagy guidelines [38] and previous studies [39, 40], several experiments were selected to monitor the drug effects on autophagy in the present study. The immunoblotting assays were conducted to explore the effects of drugs on autophagy-related protein expressions for 24-h treatment. The LC3-II/LC3-I ratio was used to measure the effects of indicated drugs on the autophagy process. The groups treated with Earle’s balanced salt solution (EBSS) for 8 h were used as positive control. To test the effects of indicated drugs on autophagy flux, the cells were treated with the indicated drugs for 24 h after pretreating with lysosome inhibitors for 1 h. The LC3-II levels normalized with β-actin were calculated for quantifying LC3II accumulation in autophagosomes. Flow cytometry with AO staining was used to measure the percentage of acidic vesicle-containing cells. MTT assays and flow cytometry with annexin V/PI double staining were respectively used to evaluate the effects of autophagy on indicated drug-mediated cytotoxicity and cell death types. Fluorescent immunocytochemistry was used to detect the colocalization of LC3 and Lamp1. All the detailed protocols for each experiment were described above.
Statistical Analysis
Statistical analyses were carried out using Sigma Plot 12.5 (Systat Software, San Diego, CA). All data are presented as the mean ± standard deviation (SD). Significant differences among groups were determined using an unpaired t test. A value of p < 0.05 was taken as an indication of statistical significance. All figures shown in this article were obtained from at least 3 independent experiments with similar results.
Results
Sunitinib Occupies an Antitumor Activity for Inducing Glioma Apoptotic Cell Death
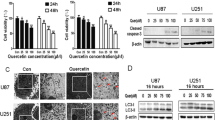
In order to reevaluate the cytotoxicity of sunitinib against gliomas, cell viability and cell death types were respectively measured in both U-87MG and A172 glioma cells using an MTT assay and annexin V-PI double staining and flow cytometric analyses. Results demonstrated that treating glioma cells with 15 or 20 μM sunitinib significantly decreased glioma cell viability (Fig. 1a, b). About 60% U-87 MG and 40% A172 cells died upon 15 μM sunitinib treatment. Therefore, we used this concentration for sunitinib treatment in the present study. In flow cytometric assays, we found that sunitinib induced apoptotic death of glioma cells (Fig. 1c, d). Furthermore, immunoblotting assays showed that sunitinib promoted caspase-3-dependent apoptosis (Fig. 1e). In addition, an additive effect on glioma cell death was observed with combined treatment with sunitinib and TMZ, a first-line treatment drug for GBM patients (Supplementary Fig. 1). Because TMZ resistance may lead to poor prognosis and recurrence of GBM patients [41], we then tested the sunitinib cytotoxicity on TMZ-resistant cells. We first established 2 TMZ-resistant cell lines, respectively named U-87-R and T98G-R (Supplementary Fig. 2). The results showed that sunitinib also possesses the ability to reduce the cell viability of TMZ-resistant glioma cells (Fig. 1f). To better understand the role of sunitinib-mediated signaling in GBM malignancy, an RNA-Seq analysis with sunitinib-treated U-87MG cells compared to parental cells was performed (Fig. 1g, h). DEGs, using the criteria of |log2FC| ≥ 1 and an FDR of ≤ 0.01, were identified, including 585 upregulated (Supplementary Table 1) and 382 downregulated genes (Supplementary Table 2). To suppose that sunitinib occupied the ability for anti-GBM tumor growth, we wondered whether sunitinib-downregulated genes contained the genes which were upregulated during GBM progression. By performing GSEAs, we found that sunitinib-downregulated genes were significantly enriched in candidates associated with GBM malignancy which were upregulated genes (multiple of change of ≥ 2 and an FDR of ≤ 0.01) between tumor and normal tissues in TCGA GBM microarray data (Fig. 1i). Results of in vitro and in silico analyses suggested that sunitinib can induce glioma cell death through repressing oncogenic signalings.
Sunitinib (St) possesses notable cytotoxicity in inducing apoptotic cell death of glioblastomas (GBMs). St mediated concentration-dependent death of both U-87 MG (a) and A172 (b) cells. (c and d) St induced apoptotic death in GBM cells. (e) St enhanced caspase-3 activation and poly(ADP ribose) polymerase (PARP) degradation. (f) St significantly enhanced cell death in temozolomide (TMZ)-resistant U-87-R and T98G-R cells. After cells were treated with the indicated dose or 15 μM St for 24 h, cell viability, cell death type, and apoptotic marker levels were respectively measured by MTT assays, annexin V-propidium iodide double staining with flow cytometric analyses, and immunoblotting. Data are the mean ± SD of 3 experiments. *p < 0.05. Relative cleavage-caspase3 (Cle-Cas3) and degradation (Deg)-PARP levels were quantified by normalizing with β-actin and showed as fold values. Heatmap (g) and volcano plot (h) showing the St-mediated transcriptome profile by RNA sequencing, including 585 upregulated and 382 downregulated genes. (i) St-downregulated genes enriched in upregulated genes of The Cancer Genome Atlas (TCGA) glioma data using gene set enrichment analyses (GSEAs)
Inhibition of Sunitinib-Induced Autophagy Potentiates Its Antitumor Activity in Glioma Cells
Due to the failure of sunitinib in treating GBM patients in clinical trials, fully understanding sunitinib-regulated signaling pathways may provide critical clues as to what is impeding the therapeutic efficacy of sunitinib. According to previous studies [42, 43], autophagy was recognized as a novel target for GBM therapy. And inhibition of autophagy enhanced sunitinib cytotoxicity in primary brain tumor initiating progenitor cell cultures (BTICs). Therefore, we aimed to identify key genes in autophagy against sunitinib in GBM. First, we utilized gene candidates that were directly or indirectly involved in autophagy in the previous literature from a human autophagy database to perform another GSEA [44]. We found that sunitinib-upregulated genes were significantly enriched in an autophagy list (Fig. 2a). To verify the effects of sunitinib in inducing autophagy in glioma cells, AO staining, flow cytometric analyses, and immunoblot assays were conducted. AO staining demonstrated that acidic vesicles had significantly increased after sunitinib treatment in both U-87 MG and A172 cells (Fig. 2b, c). Sunitinib induced LC3-II/LC3-I ratio increases and p62 degradation in a time course manner (D). Therefore, the time point of 24 h treatment was selected for monitoring sunitinib-regulating autophagy in the present study. Levels of the LC3-II/LC3-I ratio, ATG12, ATG12–ATG5 complex, and beclin-1 were increased in concentration-dependent manners after glioma cells were treated with sunitinib, whereas p62 was degraded (Fig. 2e). The groups treated with Earle’s balanced salt solution (EBSS) for 8 h were used as positive control, suggesting that sunitinib enhanced the autophagy process in GBM cells. Furthermore, sunitinib also enhanced autophagy in TMZ-resistant GBM cells (Fig. 2f). To confirm that sunitinib promoted the autophagy flux process, glioma cells were treated with sunitinib combined with CQ. CQ treatment significantly increased sunitinib-induced LC3-II accumulation which was measured by LC3-II levels normalized with β-actin expression (Fig. 2g), suggesting that sunitinib enhanced the autophagy flux in glioma cells. Next, we also found that CQ treatment enhanced sunitinib-promoted cell death (Fig. 2h, i, and Supplementary Fig. 3), suggesting that sunitinib induced protective autophagy in glioma cells. Finally, to test the effects of autophagy inhibition on sunitinib cytotoxicity, several autophagy inhibitors including HCQ, cycloheximide (CHX), and Lys05 were respectively used in combined treatment with sunitinib (Fig. 2j). The Lys05 was also selected to treat cells, identifying that sunitinib enhanced the autophagy flux in these cells by measuring LC3-II levels normalized with β-actin (Fig. 2k). We found that all combined treatments induced a more dramatic cell death compared to sunitinib treatment alone. Taken together, enhanced autophagy desensitized glioma cells to sunitinib cytotoxicity.
Inhibition of autophagy enhanced sunitinib (St) cytotoxicity in glioblastoma (GBM) cells. (a) St-upregulated genes were significantly enriched in the autophagy process by GSEAs. (b to f) St enhanced autophagy formation in both glioma cells and TMZ-resistant cells. After the cells were treated with the indicated dose or 15 μM St for 24 h or time course, autophagy signals and related marker levels were respectively detected by acridine orange staining with flow cytometric analyses and immunoblotting assays. The groups treated with Earle’s balanced salt solution (EBSS) for 8 h were used as positive control. Data are the mean ± SD of 3 experiments. *p < 0.05. (g) Chloroquine (CQ) treatment identified that St induced an autophagic flux process. Relative LC3-II, p62, ATG12, and beclin-1 levels were quantified by normalizing with β-actin and shown as fold values. After the cells were pretreated with CQ for 1 h, the sunitinib was added for another 24-h treatment. The EBSS-treated groups were used as positive control. (h to j) Respective treatment with autophagy inhibitors including CQ, hydroxychloroquine (HCQ), cycloheximide (CHX), and Lys05 enhanced St-induced cell death of GBM. (k) Lys05 treatment influenced St-mediated autophagic flux. After the cells were respectively pretreated with 10 μM CQ, 10 μM HCQ, 5 μM CHX, and 1 μM Lys05 for 1 h, St was added for another 24 h. Light chain 3 (LC3) levels, cell viability, and the apoptosis ratio were respectively measured by immunoblotting assays, MTT assays, and annexin V-propidium iodide double staining with flow cytometric analyses. Relative LC3-II levels were quantified by normalizing with β-actin and shown as fold values. Data are the mean ± SD of 3 experiments. *p < 0.05
A Survival-Associated Gene, GADD34, Was Upregulated in Sunitinib-Enhanced Autophagy of Glioma Cells
To identify critical genes involved in sunitinib-promoted autophagy, we focused on candidates that were upregulated after sunitinib treatment and those that also participated in autophagic pathways. In total, 15 gene candidates were found (Fig. 3a). We found that these genes could be categorized into several groups according to their functions, including autophagy and ER stress regulators (ATG4D, GADD34, and ERN1), gene and epigenetic regulators (SIRT1, FOXO3, FOS, and CDKN1A), heat-shock protein mediators (BAG1, BAG3, DNAJB1, and HSPB8), ubiquitination regulators (KLHL24 and SQSTM1), sphingosine phosphorylation (SPHK1), and microtubule assembly (MAP1LC3B). Among these genes, 3 candidates, viz. GADD34, CDKN1A, and SQSTM1, predicted a poor prognosis of glioma patients. By further conducting log rank tests, we identified that only high expression of GADD34 was associated with poor survival by using 3 independent databases including TCGA, Rembrandt, and GSE13041 (Fig. 3b). Next, to test the effects of sunitinib on GADD34 expression, quantitative PCR (qPCR) and immunoblotting assays were performed (Fig. 3c, d). Both messenger (m)RNA and protein levels of GADD34 increased in sunitinib-treated U-87 MG, A172, and TMZ-resistant cells. Furthermore, overexpression of GADD34 decreased sunitinib-induced cell death (Fig. 3e), whereas GADD34 depletion augmented sunitinib cytotoxicity against glioma cells (Fig. 3f and Supplementary Fig. 4). The empty pCDH and scrambled shRNA plasmids were used as control groups. We also found that the sunitinib-enhanced LC3-II/LC3-I ratio and promotion of p62 degradation were attenuated in GADD34-depleted cells (Fig. 3g). These findings suggested that GADD34 is involved in sunitinib-promoted autophagy signaling and the death of glioma cells.
GADD34 is involved in sunitinib (St)-regulated cytotoxicity and autophagy. (a) Three St-upregulated candidates were related to autophagy and risk genes in glioblastoma (GBM). HR, hazard ratio. (b) Log rank tests validated associations between 3 St-upregulated candidates and patient survival using 3 distinct databases. p < 0.05 was labeled in red color. Median survival, MS. St upregulated mRNA (c) and protein (d) levels of GADD34 in gliomas and temozolomide (TMZ)-resistant cells. Overexpression (e) and knockdown (f) of GADD34 influenced St-reduced cell viability and autophagy (g). After cells were treated with the indicated dose or 15 μM St for 24 h, cell viability, mRNA, and protein expressions were respectively measured by MTT assays, a real-time PCR, and immunoblotting assays. Data are the mean ± SD of 3 experiments. *p < 0.05. Relative GADD34 and p62 levels were quantified by normalizing with β-actin and shown as fold values. The LC3-II/LC3-I ratio was measured by evaluating the effects on the autophagy process
p38 MAPK Signaling Participates in Sunitinib-Regulated GADD34 Expression
A previous study reported that MAPK signaling is involved in GADD34 upregulation [45]. Here, we found that sunitinib-enhanced genes were significantly enriched in the MAPK pathway (Fig. 4a). Therefore, we treated glioma cells with different MAPK inhibitors, including SB203580 (a p38 MAP kinase inhibitor), SP600125 (a c-Jun N-terminal kinase 1 (JNK1), JNK2, and JNK3 inhibitor), and U0126 (a mitogen-activated protein kinase/extracellular signal-regulated kinase 1 (MEK1) and MEK2 inhibitor), to explore the roles of MAPK signaling in sunitinib-induced glioma cell death. The results demonstrated that inhibition of p38 MAPK sensitized glioma cells to sunitinib-mediated cytotoxicity (Fig. 4b). We also identified that sunitinib treatment induced an increase in p38 phosphorylation in a concentration-dependent manner (Fig. 4c). Finally, we identified that SB203580 treatment significantly abrogated sunitinib-promoted GADD34 mRNA and protein expression levels (Fig. 4d, e). Another p38 MAPK inhibitor, SB202190, was also used to confirm p38 MAPK was involved in sunitinib-mediated GADD34 gene regulation (Fig. 4f, g). No significant effects of U0126 and SP600125 on sunitinib-upregulated GADD34 expression were observed (Supplementary Fig. 5). From these findings, we confirmed that sunitinib enhanced GADD34 expression through p38 MAPK signaling.
p38-mitogen-activated protein kinase (MAPK) signaling is involved in sunitinib (St)-upregulated GADD34 expression. (a) GSEAs showed that St-upregulated genes were enriched in MAPK signaling. (b) The effects of 3 MAPK inhibitors on St-reduced cell viability. SP, SP600125; SB203, SB203580. (c) St induced p38-MAPK phosphorylation. (d to g) SB203 and SB202 (SB202190) treatment attenuated St-enhanced GADD34 mRNA and protein expressions. After cells were pretreated with 10 μM SP, SB203, SB202, or U0126 for 1 h, 15 μM St was added for another 24 h. The cell viability, and mRNA and protein expressions, were respectively measured by MTT assays, a real-time PCR, and immunoblotting assays. Data are the mean ± SD of 3 experiments. *p < 0.05. Relative p-p38 and GADD34 levels were quantified by normalizing with β-actin and shown as fold values
Guanabenz, a GADD34 Inhibitor, Enhanced Sunitinib-Mediated Cytotoxicity in Glioma Cells
Because GADD34 was identified as a crucial gene involved in sunitinib-promoted autophagy, we investigated whether its protein activity inhibitor, guanabenz, could be a potential drug to increase sunitinib antitumor effects. First, glioma cells were treated with different concentrations of guanabenz, and concentration-dependent death was observed in both U-87 MG and A172 cells (Fig. 5a). Then, 50 μM guanabenz, a noncytotoxic concentration, was selected for combining with 15 μM sunitinib treatment. Next, we found that guanabenz treatment significantly enhanced sunitinib cytotoxicity in glioma cells compared to sunitinib treatment alone (Fig. 5b). Combined treatment of glioma cells with sunitinib and guanabenz also induced more caspase-3 activation, PARP degradation, and decreased LC3-II/LC3-I ratio, suggesting that guanabenz reduced the sunitinib-induced autophagy process (Fig. 5c, d). Furthermore, by cotreatment with guanabenz and CQ, no significant LC3-II levels normalized with β-actin were increased compared with CQ treated alone (Fig. 5e, f), suggesting that guanabenz does not induce autophagy flux generation. These findings suggested that guanabenz, a GADD34 inhibitor, suppressed sunitinib-induced autophagy and increased its cytotoxicity in glioma cells.
Guanabenz (Guan), a GADD34 inhibitor, enhances sunitinib (St) efficacy in glioblastomas (GBMs). (a) Guan induced concentration-dependent cell death in GBM cells. Additive effects of combined treatment with St and Guan were observed in cell viability (b), caspase-3 activation, poly(ADP ribose) polymerase (PARP) degradation, and light chain 3 (LC3)-II accumulation (c). (d) The LC3 and Lamp1 distribution in St and Guan-treated U-87 MG cells using fluorescent immunohistochemical staining. The effects of Guan on autophagy flux by using immunoblotting assays (e) and fluorescent immunohistochemical staining (f). After cells were treated with 10 μM CQ for 1 h, 50 μM Guan was added for another 24 h. The immunoblotting assays and fluorescent immunohistochemical staining were respectively conducted. Guan treatment increased St-reduced cell viability in temozolomide (TMZ)-resistant cells (g), 1% hypoxia-cultured cells (h), sphere formation (i to k), and anchorage-independent growth ability (l). The effects of guanabenz with sunitinib on sphere formation (m) and LC3-II accumulation (n) of patient-derived GBM cells. After the cells were treated with the indicated dose of Guan alone or 50 μM Guan combined with 15 μM St, cell viability and protein expressions were respectively measured by MTT assays and immunoblotting assays. The protocols for sphere formation and soft agar assays are described in the “Materials and Methods” section. Data are the mean ± SD of 3 experiments. *p < 0.05. Relative cleavage-caspase3 (Cle-Cas3), degradation (Deg)-PARP, p62, and GADD34 levels were quantified by normalizing with β-actin and shown as fold values. The LC3-II/LC3-I ratio and LC3-II normalized with β-actin were respectively used to monitor drug effects on the autophagy process and autophagy flux
The highly aggressive nature of GBM is mainly attributed to different stress conditions such as a hypoxic microenvironment or long-term chemotherapy-induced drug resistance. Here, we wanted to explore whether guanabenz still exhibits its effects on increasing sunitinib-mediated cytotoxicity in these situations. First, we found that guanabenz enhanced sunitinib cytotoxicity on TMZ-resistant glioma cells (Fig. 5g). Similar results were also observed in 1% hypoxia-cultured glioma cells (Fig. 5h and Supplementary Fig. 6). Cancer stemness is a key feature contributing to drug resistance and tumor recurrences in GBM. Therefore, we performed a tumor sphere formation assay to investigate the combined treatment effects of sunitinib and guanabenz on glioma sphere formation. The results indicated that treating glioma cells with sunitinib or guanabenz alone significantly decreased the number and size of tumor spheres (Fig. 5i–k). Moreover, combined treatment with sunitinib and guanabenz exhibited more-significant decreases in tumor sphere numbers and sizes compared to sunitinib or guanabenz treatment alone. Soft agar assays showed that guanabenz significantly enhanced the inhibitory effect of sunitinib on anchorage-independent growth of glioma cells (Fig. 5l). The similar effects of sunitinib and guanabenz combined treatment on sphere formation and LC-3II accumulation were also observed in patient-derived HCM-BROD-0106-C71 cells (Fig. 5m, n). Taken together, we validated that inhibition of autophagy by the GADD34 inhibitor, guanabenz, sensitized glioma cells to sunitinib cytotoxicity.
Guanabenz Increase Sunitinib’s Efficacy in Suppressing GBM Growth In Vivo
To further confirm the therapeutic efficacy of the combined treatment with guanabenz and sunitinib against GBM, we performed an animal study by subcutaneously injecting U-87MG glioma cells into NOD-SCID mice. After drug treatment, we found that mice which received guanabenz or sunitinib alone exhibited decreased tumor growth compared to the control (Fig. 6a, b). More importantly, combined treatment demonstrated a more dramatic decrease in tumor growth compared to guanabenz- or sunitinib-treated mice. In addition, treatment with guanabenz, sunitinib, or their combination did not significantly lead to body weight loss compared to the control (Fig. 6c). Further detecting ki67 levels by IHC staining confirmed that combined treatment with sunitinib and guanabenz significantly decreased the proliferative rate of glioma cells in vivo (Fig. 6d). Mechanistically, we detected the autophagy marker, the LC3-II/LC3-I ratio, and p62 protein levels to understand the roles of guanabenz and sunitinib in autophagy formation. The results indicated that sunitinib induced autophagy formation in vivo, which was demonstrated by an increased LC3-II/LC3-I ratio, greater p62 protein degradation, and elevated GADD34 levels (Fig. 6e, f). Cotreatment with guanabenz blocked sunitinib-promoted autophagy, and this was also accompanied by increased cleavage of caspase-3 suggesting more apoptotic cell death in the combined treatment group. From these findings, we confirmed that guanabenz blocked sunitinib-promoted autophagy and enhanced antitumor activity of sunitinib in a GBM xenograft model.
Guanabenz (Guan) promoted sunitinib (St)-suppressing tumor growth in vivo. The effects of the combined treatment with St and Guan were measured in tumor growth (a), tumor volume (b), body weight (c), Ki67 levels by immunochemistry staining (d), and autophagy-related marker expression by immunoblotting assays (e). (f) Quantitative results were measured from immunoblot data in (e). Each group contained 5 subjects. Twenty percent dimethyl sulfoxide (DMSO) was used as the vehicle control. The detailed protocol for xenograft studies is described in the “Materials and Methods” section. The red arrow markers indicate the drug injection date. *p < 0.05
Discussion
In response to cytotoxic drug treatment, malignant tumors are able to trigger different mechanisms to survive, and one of these mechanisms is autophagy. In our study, sunitinib promoted autophagy signaling in glioma cells. Through transcriptome screening by RNA-Seq, we identified key genes involved in sunitinib-regulated autophagy. By analyzing associations of these sunitinib-regulated autophagy genes with GBM patient survival, elevated GADD34 expression was strongly associated with a poor prognosis in multiple GBM patient cohorts. GADD34 is involved in sunitinib-mediated autophagy through p38-MAPK signaling. Furthermore, guanabenz, a GADD34 activity inhibitor, enhanced sunitinib cytotoxicity and abrogated sunitinib-induced autophagy against glioma cells. Using 1% hypoxia-cultured cells, TMZ-resistant cells, glioma stem cells, and an in vivo study, we confirmed that the combination of guanabenz and sunitinib significantly increased sunitinib-promoted glioma cell death, decreased sunitinib-promoted autophagy, and enhanced sunitinib-reduced tumor growth. Taken together, our findings identified that autophagy signaling plays a crucial role in protecting against sunitinib-mediated GBM suppression, and GADD34 is an important mediator involved in this process. Our results also suggested that guanabenz may be a good adjuvant drug for enhancing sunitinib cytotoxicity in GBM.
The role of autophagy in tumor progression is not universal. By removing damaged organelles, autophagy promotes tumor cell survival [46,47,48]. On the other hand, autophagy also induces cell death through eliciting self-eating [49,50,51]. In GBM, several studies reported that resistance to clinically used drugs such as TMZ [52] and bevacizumab [31] is linked to autophagy formation. Herein, we found that sunitinib, a multiple TKI, promotes protective autophagy in GBM. This finding suggested that autophagy is also involved in the poor efficacy of sunitinib in GBM. Different studies have reported conflicting mechanisms of sunitinib in autophagy induction. In renal clear cell carcinoma, it was found that sunitinib, a hydrophobic weak base drug, could accumulate in lysosomes and interrupt their function [21]. This further leads to impairment of autolysosome formation and inhibition of autophagic flux. In contrast, it was found that combined treatment with sunitinib and lys05, an autophagy inhibitor, influenced the LC3-II accumulation and p62 levels compared to sunitinib or lys05 treatment alone in ovarian cancer [23], indicating that sunitinib induces autophagy flux formation. Therefore, the diverse mechanisms of sunitinib-induced autophagy might be dependent on the cancer type. In our study, with CQ and sunitinib cotreatment, we identified autophagic flux induction by sunitinib in GBM. Suppression of autophagy using different autophagy inhibitors enhanced sunitinib-mediated glioma suppression. Hence, blocking of autophagy induction is an attractive approach to enhance the therapeutic potential of sunitinib in GBM.
GADD34 is a member of gadd and MyD mammalian genes encoding acidic proteins which were originally found to possess the function of suppressing cell growth [53]. GADD34 was reported to be upregulated by amino acid deprivation or endoplasmic reticular (ER) stress [54]. During ER stress, the activated unfolded protein response (UPR) induces PKR-like ER kinase (PERK)-controlled signaling. Although PERK-regulated pathways mainly suppress protein translation, 2 downstream targets, GADD34 and CHOP, are upregulated [55]. Moreover, silencing of GADD34 during ER stress leads to premature cell death [35]. This finding implies a protective role of GADD34 against ER stress-promoted cell death. As for the mechanisms of GADD34 in protecting cells from apoptosis, it was found that upregulation of GADD34 expression levels is accompanied by mTOR inactivation and autophagy promotion during ER stress [35]. Moreover, another study utilized GADD34-knockout mice to demonstrate that GADD34 suppresses mTOR activity through dephosphorylating p-TSC2 and initiates autophagy during starvation [34]. Those studies suggested an important role of GADD34 in inducing protective autophagy under stress conditions. Nonetheless, few studies have investigated the role of GADD34-mediated autophagy in cancers. Herein, we demonstrated that GADD34 predicted a poor prognosis in multiple glioma patient cohorts implying that GADD34 might be an important molecule in regulating GBM malignancy. Through a genome-wide transcriptome analysis, we identified that sunitinib upregulated GADD34 gene expression. Mechanistically, we found that GADD34 is involved in sunitinib-induced autophagy, and depletion of GADD34 sensitized glioma cells to sunitinib treatment. Taken together, these results imply that GADD34 may be a potential therapeutic target for GBM.
Guanabenz, an alpha agonist of the alpha-2 adrenergic receptor, is an FDA-approved antihypertension drug [56]. It was reported that in addition to its function as an alpha-2 adrenergic receptor agonist, guanabenz is also able to inhibit GADD34-mediated downstream protein dephosphorylation [57]. It was also found that thapsigargin-induced autophagy was attenuated by guanabenz treatment [35]. Therefore, in our present study, guanabenz was suggested to be a potential drug that could be evaluated for blocking sunitinib-promoted autophagy in gliomas. We performed both in vitro and in vivo experiments to show that combined treatment with guanabenz and sunitinib suppressed cell viability, anchorage-dependent growth, sphere formation, and in vivo tumor growth of glioma cells more efficiently than did sunitinib alone. Furthermore, we found that this combined treatment also occupied better cytotoxicity for inducing TMZ-resistant cell death. Because TMZ resistance may lead to poor prognosis and recurrence of GBM patients, we suggest that such drugs like guanabenz in inhibiting GADD34 functions or expression may be potential adjuvant drugs with sunitinib for GBM therapy.
In the present study, we established a sunitinib-mediated gene signature by using RNA seq. We found that in addition to GADD34, other 14 sunitinib-upregulated genes were all recognized as autophagy-related genes. These genes respectively belonged to distinct signaling pathways, including heat-shock protein regulation, ubiquitination, sphingosine phosphorylation, and microtubule assembly. By literature searching, sunitinib has been reported to induce ubiquitination in regulating insulin-like growth factor receptor subtype 1 (IGF-1R) expression [58]. Sphingosine kinase-1 activation was observed in renal cell carcinoma cells for developing acquired resistance against sunitinib [59]. However, the detailed mechanisms to connect these signalings and sunitinib resistance with autophagy are still unclear, especially the heat-shock proteins. These need further investigations in the future. There are still some limitations in the present study. For example, guanabenz is not a specific drug for targeting GADD34. A better directly targeting drug to GADD34 is needed for combined treatment with sunitinib. Other unknown genes and mechanisms are still needed to be explored for understanding the development of acquired resistance against sunitinib. In addition, although previous studies identified that intravenous guanabenz [56, 60] and sunitinib [61] can penetrate the blood–brain barrier (BBB) by using in vivo models, the degrees of these 2 drug penetrations through BBB were limited. Using the spontaneous brain tumor-forming or intracranial tumor implantation animal models may be more accurate to evaluate the drug efficacy. Furthermore, to develop strategies for improving the penetration of drugs across the BBB is a critical issue for future GBM therapy. Consequentially, we identified that sunitinib induced protective autophagy through upregulating GADD34 expression in GBM. Exploring a GADD34-targeting drug such as guanabenz can guide new therapeutic approaches to enhance the efficacy of sunitinib in GBM.
References
Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016;131(6):803-820.
Louis DN, Perry A, Burger P, et al. International Society Of Neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 2014;24(5):429-435.
Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin 2010;60(3):166-193.
Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009;10(5):459-66.
Fleming TP, Saxena A, Clark WC, et al. Amplification and/or overexpression of platelet-derived growth factor receptors and epidermal growth factor receptor in human glial tumors. Cancer Res 1992;52(16):4550-4553.
Folkins C, Shaked Y, Man S, et al. Glioma tumor stem-like cells promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1. Cancer Res 2009;69(18):7243-7251.
Popescu AM, Alexandru O, Brindusa C, et al. Targeting the VEGF and PDGF signaling pathway in glioblastoma treatment. Int J Clin Exp Pathol 2015;8(7):7825-7837.
Reardon DA, Egorin MJ, Desjardins A, et al. Phase I pharmacokinetic study of the vascular endothelial growth factor receptor tyrosine kinase inhibitor vatalanib (PTK787) plus imatinib and hydroxyurea for malignant glioma. Cancer 2009;115(10):2188-2198.
Batchelor TT, Sorensen AG, di Tomaso E, et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell 2007;11(1):83-95.
Balaña C, Gil MJ, Perez P, et al. Sunitinib administered prior to radiotherapy in patients with non-resectable glioblastoma: results of a phase II study. Target Oncol 2014;9(4):321-329.
Hatipoglu G, Hock SW, Weiss R, et al. Sunitinib impedes brain tumor progression and reduces tumor-induced neurodegeneration in the microenvironment. Cancer Sci 2015;106(2):160-170.
Giannopoulou E, Dimitropoulos K, Argyriou AA, Koutras AK, Dimitrakopoulos F, Kalofonos HP. An in vitro study, evaluating the effect of sunitinib and/or lapatinib on two glioma cell lines. Investig New Drugs 2010;28(5):554-560.
de Bouard S, Herlin P, Christensen JG, et al. Antiangiogenic and anti-invasive effects of sunitinib on experimental human glioblastoma. Neuro-Oncology 2007;9(4):412-423.
Martinho O, Silva-Oliveira R, Miranda-Gonçalves V, et al. In vitro and in vivo analysis of RTK inhibitor efficacy and identification of its novel targets in glioblastomas. Transl Oncol 2013;6(2):187-196.
Chahal M, Xu Y, Lesniak D, et al. MGMT modulates glioblastoma angiogenesis and response to the tyrosine kinase inhibitor sunitinib. Neuro-oncology 2010;12(8):822-833.
Martinho O, Zucca LE, Reis RM. AXL as a modulator of sunitinib response in glioblastoma cell lines. Exp Cell Res 2015;332(1):1-10.
Neyns B, Sadones J, Chaskis C, et al. Phase II study of sunitinib malate in patients with recurrent high-grade glioma. J Neuro-Oncol 2011;103(3):491-501.
Wetmore C, Daryani VM, Billups CA, et al. Phase II evaluation of sunitinib in the treatment of recurrent or refractory high-grade glioma or ependymoma in children: a children’s Oncology Group Study ACNS1021. Cancer Med 2016;5(7):1416-1424.
Pan E, Yu D, Yue B, et al. A prospective phase II single-institution trial of sunitinib for recurrent malignant glioma. J Neuro-Oncol 2012;110(1):111-118.
Reardon DA, Vredenburgh JJ, Coan A, et al. Phase I study of sunitinib and irinotecan for patients with recurrent malignant glioma. J Neuro-Oncol 2011;105(3):621-627.
Giuliano S, Cormerais Y, Dufies M, et al. Resistance to sunitinib in renal clear cell carcinoma results from sequestration in lysosomes and inhibition of the autophagic flux. Autophagy 2015;11(10):1891-1904.
Wang B, Lu D, Xuan M, Hu W. Antitumor effect of sunitinib in human prostate cancer cells functions via autophagy. Exp Ther Med 2017;13(4):1285-1294.
DeVorkin L, Hattersley M, Kim P, et al. Autophagy inhibition enhances sunitinib efficacy in clear cell ovarian carcinoma. Mol Cancer Res 2017;15(3):250-258.
Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol 2010;221(1):3-12.
Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 2011;10(9):1533-1541.
Li ZY, Yang Y, Ming M, Liu B. Mitochondrial ROS generation for regulation of autophagic pathways in cancer. Biochem Biophys Res Commun 2011;414(1):5-8.
Ramakrishnan S, Nguyen TM, Subramanian IV, Kelekar A. Autophagy and angiogenesis inhibition. Autophagy 2007;3(5):512-515.
Li B, Zhou C, Yi L, Xu L, Xu M. Effect and molecular mechanism of mTOR inhibitor rapamycin on temozolomide-induced autophagic death of U251 glioma cells. Oncol Lett 2018;15(2):2477-2484.
Huang H, Song J, Liu Z, Pan L, Xu G. Autophagy activation promotes bevacizumab resistance in glioblastoma by suppressing Akt/mTOR signaling pathway. Oncol Lett 2018;15(2):1487-1494.
Golden EB, Cho HY, Jahanian A, et al. Chloroquine enhances temozolomide cytotoxicity in malignant gliomas by blocking autophagy. Neurosurg Focus 2014;37(6):E12.
Abdul Rahim SA, Dirkse A, Oudin A, et al. Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A. Br J Cancer 2017;117(6):813-825.
Wiedmer T, Blank A, Pantasis S, et al. Autophagy inhibition improves sunitinib efficacy in pancreatic neuroendocrine tumors via a lysosome-dependent mechanism. Mol Cancer Ther 2017;16(11):2502-2515.
Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol 2003;23(4):1292-1303.
Uddin MN, Ito S, Nishio N, Suganya T, Isobe K. Gadd34 induces autophagy through the suppression of the mTOR pathway during starvation. Biochem Biophys Res Commun 2011;407(4):692-698.
Holczer M, Banhegyi G, Kapuy O. GADD34 keeps the mTOR pathway inactivated in endoplasmic reticulum stress related autophagy. PLoS One 2016;11(12):e0168359.
Zhou Q, Gallo JM. Differential effect of sunitinib on the distribution of temozolomide in an orthotopic glioma model. Neuro-oncology 2009;11(3):301-310.
Konrad C, Queener SF, Wek RC, Sullivan WJ, Jr. Inhibitors of eIF2α dephosphorylation slow replication and stabilize latency in Toxoplasma gondii. Antimicrob Agents Chemother 2013;57(4):1815-1822.
Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12(1):1-222.
Lin CJ, Lee CC, Shih YL, et al. Inhibition of mitochondria- and endoplasmic reticulum stress-mediated autophagy augments temozolomide-induced apoptosis in glioma cells. PLoS One 2012;7(6):e38706.
Lin CJ, Lee CC, Shih YL, et al. Resveratrol enhances the therapeutic effect of temozolomide against malignant glioma in vitro and in vivo by inhibiting autophagy. Free Radic Biol Med 2012;52(2):377-391.
Jiapaer S, Furuta T, Tanaka S, Kitabayashi T, Nakada M. Potential strategies overcoming the temozolomide resistance for glioblastoma. Neurol Med Chir (Tokyo) 2018;58(10):405-421.
Kaza N, Kohli L, Roth KA. Autophagy in brain tumors: a new target for therapeutic intervention. Brain Pathol 2012;22(1):89-98.
Moeckel S, LaFrance K, Wetsch J, et al. ATF4 contributes to autophagy and survival in sunitinib treated brain tumor initiating cells (BTICs). Oncotarget 2019;10(3):368-382.
Moussay E, Kaoma T, Baginska J, et al. The acquisition of resistance to TNFalpha in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy 2011;7(7):760-770.
Oh-Hashi K, Maruyama W, Isobe K. Peroxynitrite induces GADD34, 45, and 153 VIA p38 MAPK in human neuroblastoma SH-SY5Y cells. Free Radic Biol Med 2001;30(2):213-221.
Wu Z, Chang PC, Yang JC, et al. Autophagy blockade sensitizes prostate cancer cells towards Src family kinase inhibitors. Genes Cancer 2010;1(1):40-49.
O'Donovan TR, O'Sullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy 2011;7(5):509-524.
Natsumeda M, Aoki H, Miyahara H, et al. Induction of autophagy in temozolomide treated malignant gliomas. Neuropathology 2011;31(5):486-493.
Yu L, Wan F, Dutta S, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A 2006;103(13):4952-4957.
Yu L, Alva A, Su H, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 2004;304(5676):1500-1502.
Shimizu S, Kanaseki T, Mizushima N, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 2004;6(12):1221-1228.
Knizhnik AV, Roos WP, Nikolova T, et al. Survival and death strategies in glioma cells: autophagy, senescence and apoptosis triggered by a single type of temozolomide-induced DNA damage. PLoS One 2013;8(1):e55665.
Zhan Q, Lord KA, Alamo I, Jr., et al. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol Cell Biol 1994;14(4):2361-2371.
Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta 2013;1833(12):3460-3470.
Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem 2003;278(37):34864-34873.
Holmes B, Brogden RN, Heel RC, Speight TM, Avery GS. Guanabenz. A review of its pharmacodynamic properties and therapeutic efficacy in hypertension. Drugs 1983;26(3):212-229.
Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 2011;332(6025):91-94.
Shen H, Fang Y, Dong W, Mu X, Liu Q, Du J. IGF-1 receptor is down-regulated by sunitinib induces MDM2-dependent ubiquitination. FEBS Open Bio 2012;2:1-5.
Gao H, Deng L. Sphingosine kinase-1 activation causes acquired resistance against sunitinib in renal cell carcinoma cells. Cell Biochem Biophys 2014;68(2):419-425.
Baum T, Shropshire AT. Studies on the centrally mediated hypotensive activity of guanabenz. Eur J Pharmacol 1976;37(1):31-44.
Tang SC, Lagas JS, Lankheet NA, et al. Brain accumulation of sunitinib is restricted by P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) and can be enhanced by oral elacridar and sunitinib coadministration. Int J Cancer 2012;130(1):223-233.
Acknowledgments
This study was sponsored by grants from the Ministry of Science and Technology, Taiwan (MOST 106-2320-B-038-051-MY3, MOST 108-2314-B-038-110, and MOST 109-2320-B-038-014). We thank the National RNAi Core Facility at Academia Sinica, Taiwan, for providing shRNA reagents and related services, and used models and data derived by the Human Cancer Models Initiative (HCMI); dbGaP accession number phs001486. We would also like to acknowledge the Laboratory Animal Center at Taipei Medical University for the in vivo xenograft experiments. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ho, KH., Lee, YT., Chen, PH. et al. Guanabenz Sensitizes Glioblastoma Cells to Sunitinib by Inhibiting GADD34-Mediated Autophagic Signaling. Neurotherapeutics 18, 1371–1392 (2021). https://doi.org/10.1007/s13311-020-00961-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13311-020-00961-z