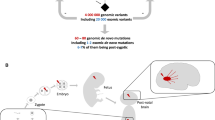
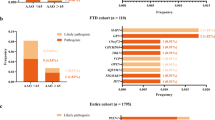
Abstract
Purpose of review
Methods for the genetic diagnosis of neurodegenerative disorders were reviewed, including their backgrounds and applications in the laboratory. Majority of disease-causing gene mutations were uncommon in the general population, where dominant variations could be easily identified in certain disorders. The development of molecular, next generation sequencing (NGS) and cytogenetic techniques allowed to identify multiple genetic mutations leading to diseases. Using of the accurate multivariate diagnosis of diseases would be essential for appropriate treatment of patients, genetic counseling and prevention strategies
Recent findings
Abnormal genes play an extremely important role in the pathogenesis of neurodegenerative disorders of the nervous system. Many studies of genetic have clearly indicated that the molecular mechanisms underlying the etiology and pathogenesis of most neurodegenerative disorders until now. Mutation is necessary for life, while fidelity is necessary for DNA replication. Mistakes in DNA replication/transcription can cause cells to produce either too much or too little protein or a defective protein. Studies on mutations have revealed the normal functions of genes, messages, proteins, the causes of many diseases, and the variability of responses among individuals. Based on the presence of damaging variants, there are many genes have identified that associated with neurodegenerative disorders through to genetic analyses of patients. This declaration is interesting because also our previous analysis has shown that several types of neurodegenerative diseases associated with abnormal genes function have been well described, including Alzheimer’s disease (AD), front temporal dementia (FTD), amyotrophic lateral sclerosis (ALS), prion disease, and Parkinson’s disease (PD). Since the potential treatment strategies for these disorders may be more successful during the pre-clinical stages than in the actual clinical setup, accurate, simple, and affordable diagnostic methods are needed. A mutation can be defined as a sequence change in a test sample compared to the sequence of a reference standard. This has led to the development of methods for screening and detecting abnormal DNA, including techniques that detect previously described mutations (genotyping) and those that scan for mutations in a particular target region (mutation scanning). This review provides a broad overview of the range of currently available mutation detection techniques with special emphasis on neurodegenerative disorders in humans. We have discussed the methods used for detecting unknown mutations, such as ribonuclease, denaturing gradient-gel electrophoresis, carbodiimide, chemical cleavage, single- strand conformation polymorphism, heteroduplex, and sequencing. Furthermore, other diagnostic methods for testing mutations include allele-specific oligonucleotide hybridization; allele-specific amplification, ligation, primer extension, and the artificial introduction of restriction sites are also reviewed. In order to identify the mutation, the last of the work provides basic insights into some of the most popular in silico tools for splicing defect prediction from the viewpoint of end users.
Similar content being viewed by others
References
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006).
Schork, N. J., Fallin, D. & Lanchbury, J. S. Single nucleotide polymorphisms and the future of genetic epidemiology. Clinical Genetics 58, 250–264 (2000).
Kruglyak, L. & Nickerson, D. A. Variation is the spice of life. Nature Genet 27, 234–236 (2001).
Stephens, J. C. et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science 293, 489–493 (2001).
Bromberg, Y. & Rost, B. SNAP: predict effect of nonsynonymous polymorphisms on function. Nucleic Acids Res 35, 3823–3835 (2007).
Emahazion, T. et al. SNP association studies in Alzheimer’s disease highlight problems for complex disease analysis. Trends in Genetics 17, 407–413 (2001).
Hattersley, A. T. & McCarthy, M. I. What makes a good genetic association study? Lancet 366, 1315–1323 (2005).
Iafrate, A. J. et al. Detection of large-scale variation in the human genome. Nat Genet 36, 949–951 (2004).
Sharp, A. J. et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77, 78–88 (2005).
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nat Genet 37, 727–732 (2005).
Dean, M. Approaches to identify genes for complex human diseases: Lessons from Mendelian disorders. Hum Mutat 22, 261–274 (2003).
Mathew, C. Science medicine and the future: Postgenomic technologies: hunting the genes for common disorders. BMJ 322, 1031–1034 (2001).
Cardon, L. R. & Bell, J. I. Association study designs for complex diseases. Nat Rev Genet 2, 91–99 (2001).
Giau, V. V., Bagyinszky, E., An, S. S. & Kim, S. Y. Role of apolipoprotein E in neurodegenerative diseases. Neuropsychiatr Disease Treatment 11, 1723–1737 (2015).
Mathew, C. G. DNA diagnostics: goals and Challenges. Br Med Bull 55, 325–339 (1999).
Giau, V. V., An, S. S., Bagyinszky, E. & Kim, S. Y. Gene panels and primers for next generation sequencing studies on neurodegenerative disorders. Molecular & Cellular Toxicology 11, 89–143 (2015).
Hebert, L. E., Scherr, P. A., Bienias, J. L., Bennett, D. A. & Evans, D. A. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol 60, 1119–1122 (2003).
Bird. D. T. Genetic Aspects of Alzheimer Disease. Genet Med 10, 231–239 (2008).
Knopman, D. S. et al. Practice parameter: diagnosis of dementia (an evidence-based review). Report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology 56, 1143–1153 (2001).
Perl, D. P. Neuropathology of Alzheimer’s disease. Mt Sinai J Med 77, 32–42 (2010).
Sun, Y. et al. Next-generation diagnostics: gene panel, exome, or whole genome? Hum Mutat 36, 648–655 (2015).
Meyer, J. M. & Breitner, J. C. Multiple threshold model for the onset of Alzheimer’s disease in the NAS-NRC twin panel. Am J Med Genet 81, 92–97 (1998).
Rossor, M. N., Fox, N. C., Mummery, C. J., Schott, J. M. & Warren, J. D. The diagnosis of young-onset dementia. Lancet Neurol 9, 793–806 (2010).
Rogaev, E. I. et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature 376, 775–778 (1995).
Levy-Lahad, E. et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 269, 973–977 (1995).
Sherrington, R. et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 375, 754–760 (1995).
Kowalska, A. et al. Genetic study of familial cases of Alzheimer’s disease. Acta Biochim Pol 51, 245–252 (2004).
Rossor, M. N. et al. The diagnosis of young-onset dementia. Lancet Neurol 9, 793–806 (2010).
Tanzi, R. E. et al. Assessment of amyloid β-protein precursor gene mutations in a large set of familial and sporadic Alzheimer disease cases. Am J Med Genet 51, 273–282 (1992).
Gatz, M. et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 63, 168–174 (2006).
Selkoe, D. J. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev 81, 741–766 (2001).
Barnes, D. E. & Yaffe, K. The projected effect of risk factor reduction on Alzheimer’s disease prevalence. Lancet Neurol 10, 819–828 (2011).
Strittmatter, W. J. et al. Apolipoprotein E: highavidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA 90, 1977–1981 (1993).
Namba, Y., Tamonaga, M., Kawasaki, H., Otomo, E. & Ikeda, K. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and cru plaque amyloid in Creutzfeldt-Jakob disease. Brain Research 541, 163–166 (1991).
Pericak-Vance, M. A. et al. Linkage studies in familial Alzheimer’s disease: evidence for chromosome 19 linkage. Am J Hum Genet 48, 1034–1050 (1991).
Corder, E. H. et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Gene 7, 180–184 (1994).
Farrer, L. A. et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. JAMA 278, 1349–1356 (1997).
Locke, P. A., Conneally, P. M., Tanzi, R. E., Gusella, J. F. & Haines, J. L. Apolipoprotein E4 allele and Alzheimer disease: examination of allelic association and effect on age at onset in both early-and late-onset cases. Genet Epidemiol 12, 83–92 (1995).
Bergem, A. L. Heredity in dementia of the Alzheimer type. Clin Genet 46, 144–149 (1994).
Rabbani, B., Tekin, M. & Mahdieh, N. The promise of whole-exome sequencing in medical genetics. J Human Genetics 59, 5–15 (2014).
So, H. C., Gui, A. H., Cherny, S. S. & Sham, P. C. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol 35, 310–317 (2011).
Grupe, A. et al. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum Mol Genet 16, 865–873 (2007).
Coon, K. D. et al. A highdensity whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry 68, 613–618 (2007).
Corder, E. H. et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 7, 180–184 (1994).
Lambert, J. C. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41, 1094–1099 (2009).
Harold, D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41, 1088–1093 (2009).
Seshadri, S. et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303, 1832–1840 (2010).
Naj, A. C. et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with lateonset Alzheimer’s disease. Nat Genet 43, 436–441 (2011).
Hollingworth, P. et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43, 429–435 (2011).
Rogaeva, E. et al. The neuronal sortilinrelated receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet 39, 168–177 (2007).
Jonsson, T. et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 368, 107–116 (2013).
Lambert, J. C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45, 1452–1458 (2013).
Tosto, G. et al. F-box/LRR-repeat protein 7 is genetically associated with Alzheimer’s disease. Ann Clin Transl Neurol 2, 810–820 (2015).
Kim, M. et al. Potential late-onset Alzheimer’s disease-associated mutations in the ADAM10 gene attenuate {alpha}-secretase activity. Hum Mol Genet 18, 3987–3996 (2009).
Cruchaga, C. et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature 505, 550–554 (2014).
Bird, D. T. Genetic Aspects of Alzheimer Disease. Genet Med 10, 231–239 (2008).
Logue, M. W. et al. Two rare AKAP9 variants are associated with Alzheimer’s disease in African Americans. Alzheimers Dement 10, 609–618.e11 (2014).
Allen, M. et al. Late-onset Alzheimer disease risk variants mark brain regulatory loci. Neurol Genet 1, e15 (2015).
Plotkin, J. B. & Kudla, G. Synonymous but not the same: the causes and consequences of codon bias. Nature Rev Genet 12, 32–42 (2011).
Chamary, J. V. & Hurst, L. D. The price of silent mutations. Scientific American 300, 46–53 (2009).
Schattner, P. & Diekhans, M. Regions of extreme synonymous codon selection in mammalian genes. Nucleic Acids Res 34, 1700–1710 (2006).
Kimchi-Sarfaty, C. et al. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528 (2007).
Sauna, Z. E. & Kimchi-Sarfaty, C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12, 683–691 (2011).
Chen, R., Davydov, E. V., Sirota, M. & Butte, A. J. Nonsynonymous and synonymous coding SNPs show similar likelihood and effect size of human disease association. PLoSONE 5, e13574 (2010).
Nyrén, P. Enzymatic method for continuous monitoring of DNA polymerase activity. Analytical Biochemistry 167, 235–248 (1987).
Greenwood, P. M., Lin, M. K., Sundararajan, R., Fryxell, K. J. & Parasuraman, R. Synergistic effects of genetic variation in nicotinic and muscarinic receptors on visual attention but not working memory. Proc Natl Acad Sci USA 106, 3633–3638 (2009).
Lu, J. et al. Polymorphic Variation in Cytochrome Oxidase Subunit Genes. J Alzheimers Dis 21, 141–154 (2010).
Yang, S. Y., He, X. Y. & Miller, D. D. HSD17B10: A gene involved in cognitive function through metabolism of isoleucine and neuroactive steroids. Mol Genet Metab 92, 36–42 (2007).
Ribases, M. et al. Association study of 10 genes encoding neurotrophic factors and their receptors in adult and child attention-deficit/hyperactivity disorder. Biol Psychiatr 63, 935–945 (2008).
Dhaenens, C. M. et al. A genetic variation in the ADORA2A gene modifies age at onset in Huntington’s disease. Neurobiol Dis 35, 474–476 (2009).
Watanabe, Y. et al. A two-stage case-control association study of PADI2 with schizophrenia. J Human Genetics 54, 430–432 (2009).
Sebat, J. et al. Large-Scale Copy Number Polymorphism in the Human Genome. Science 305, 525–528 (2004).
Tzvetkov, M. V., Brockmoller, J., Roots, I. & Kirchheiner, J. Common genetic variations in human brain-specific tryptophan hydroxylase-2 and response to antidepressant treatment. Pharmacogenet Genomics 18, 495–506 (2008).
Zhou, Q. et al. Detection of RASA1 mutations in patients with sporadic Sturge-Weber syndrome. Childs Nerv Syst 27, 603–607 (2011).
Ho, P. Y. et al. Simultaneous assessment of the effects of exonic mutations on RNA splicing and protein functions. Biochem Biophys Res Commun 373, 515–520 (2008).
Nackley, A. G. et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science 314, 1930–1933 (2006).
Oberacher, H. et al. Direct molecular haplotyping of multiple polymorphisms within exon 4 of the human catechol-Omethyltransferase gene by liquid chromatography-electrospray ionization time-of-flight mass spectrometry. Anal Bioanal Chem 386, 83–91 (2006).
Orita, M., Iwahana, H., Kanazawa, H., Hayashi, K. & Sekiya, T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A 86, 2766–2770 (1989).
Sarmiento, U. M. & Storb, R. F. Restriction fragment length polymorphism of the major histocompatibility complex of the dog. Immunogenetics 28, 117–124 (1988).
Rosenbaum, V. & Riesner, D. Temperature-gradient gel electrophoresis. Thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extract. Biophys Chem 26, 235–246 (1987).
Wartell, R. M., Hosseini, S. H. & Moran, C. P. Detecting base pair substitutions in DNA fragments by temperature-gradient gel electrophoresis. Nucleic Acids Res 18, 2699–2705 (1990).
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74, 5463–5467 (1977).
Ronaghi, M. Pyrosequencing sheds light on DNA sequencing. Genome Research 11, 3–11 (2011).
Ronaghi, M. et al. Real-time DNA sequencing using detection of pyrophosphate release. Analytical Biochemistry 242, 84–89 (1996).
Ronaghi, M. Improved performance of pyrosequencing using single-stranded DNA-binding protein. Analytical Biochemistry 286, 282–288 (2000).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–480 (2005).
Rabbani, B., Tekin, M. & Mahdieh, N. The promise of whole-exome sequencing in medical genetics. J Human Genetics 59, 5–15 (2014).
Botstein, D., White, R. L., Skolnick, M. & David, R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Human Genetics 32, 314–331 (1980).
Lyamichev, V. et al. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nature Biotech 17, 292–296 (1999).
Hall, J. G. et al. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc Natl Acad Sci USA 97, 8272–8277 (2000).
McGall, G. H. & Christians, F. C. High-density GeneChip oligonucleotide probe arrays. Adv Biochem Eng Biotechnol 77, 21–42 (2002).
Landegren, U., Kaiser, R., Sanders, J. & Hood, L. A ligase-mediated gene detection technique. Science 1988, 241, 1077–1080.
Kim, S. & Misra, A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng 9, 289–320 (2007).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249 (2010).
Bao, L., Zhou, M. & Cui, Y. nsSNPAnalyzer: Identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res 33, 480–482 (2005).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42, 252–258 (2014).
Tavtigian, S. V., Greenblatt, M. S., Lesueur, F. & Byrnes, G. B. In silico analysis of missense substitutions using sequence-alignment based methods. Hum Mutat 29, 1327–1336 (2008).
Leong, I. U., Skinner, J. & Love, D. Application of massively parallel sequencing in the clinical diagnostic testing of inherited cardiac conditions. Med Sci 2, 98–126 (2014).
Bromberg, Y. & Rost, B. SNAP: predict effect of nonsynonymous polymorphisms on function. Nucleic Acids Res 35, 3823–3835 (2007).
Calabrese, R., Capriotti, E., Fariselli, P., Martelli, P. L. & Casadio, R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat 30, 1237–1244 (2009).
Ng, P. C. & Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res 11, 863–874 (2001).
Ng, P. C. & Henikoff, S. Accounting for human polymorphisms predicted to affect protein function. Genome Res 12, 436–446 (2002).
Choi, Y., Sims, G. E., Murphy, S., Miller, J. R. & Chan, A. P. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7, e46688 (2012).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymouse variants on protein function using the SIFT algorithm. Nat Protocols 4, 1073–1081 (2009).
Mi, H., Muruganujan, A. & Thomas, P. D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41, 377–386. doi:10.1093/nar/gks1118 (2013).
Stone, E. A. & Sidow, A. Physicochemical constraint violation by missense substitution mediates impairment of protein function and disease severity. Genome Res 15, 978–986 (2005).
Carmel, I., Tal, S., Vig, I. & Ast, G. Comparative analysis detects dependencies among the 5ʹ splice-site positions. RNA 10, 828–840 (2004).
Shapiro, M. B. & Senapathy, P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15, 7155–7174 (1987).
Reese, M. G., Eeckman, F. H., Kulp, D. & Haussler, D. Improved splice site detection in Genie. J Comput Biol 4, 311–323 (1997).
Pertea, M., Lin, X. & Salzberg, S. L. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res 29, 1185–1190 (2001).
Desmet, F. O. et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37, e67 (2009).
Reva, B., Antipin, Y. & Sander, C. Predicting the functional impact of protein mutations: Applications to cancer genomics. Nucleic Acids Res 39, e118 (2011).
Dogan, R. I., Getoor, L., Wilbur, W. J. & Mount, S. M. SplicePort: an interactive splice-site analysis tool. Nucleic Acids Res 35, 285–291 (2007).
Ryan, M., Diekhans, M., Lien, S., Liu, Y. & Karchin, R. LS-SNP/PDB: Annotated non-synonymous SNPs mapped to protein data bank structures. Bioinformatics 25, 1431–1432 (2009).
De Baets, G. et al. SNP effect 4.0: Online prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res 40, 935–939 (2012).
Capriotti, E., Fariselli, P. & Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33, 306–310 (2005).
Källberg, M. et al. Template-based protein structure modeling using the RaptorX web server. Nature Protocols 7, 1511–1522 (2012).
Webb, B. & Sali, A. Comparative Protein Structure Modeling Using Modeller. Curr Protoc Bioinformatics 47, 5.6.1–5.6.32. doi:10.1002/0471250953.bi0506s47 (2014).
Yang, J. et al. The I-TASSER Suite: Protein structure and function prediction. Nature Methods 12, 7–8. doi:10.1038/nmeth.3213 (2015).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10, 845–858 (2015).
Belmont, J. W. & Leal, S. M. Complex phenotypes and complex genetics: an introduction to genetic studies of complex traits. Curr Atheroscler Rep 27, 180–187 (2005).
Cardon, L. R. & Bell, J. I. Association study designs for complex diseases. Nat Rev Genet 2, 91–99 (2001).
Stone, E. A. & Sidow, A. Physicochemical constraint violation by missense substitution mediates impairment of protein function and disease severity. Genome Res 15, 978–986 (2005).
van Dijk, E. L., Auger, H., Jaszczyszyn, Y. & Thermes, C. Ten years of next-generation sequencing technology. Trends Genet 30, 418–426 (2014).
Wade, C. H., Tarini, B. A. & Wilfond, B. S. Growing up in the genomic era: implications of whole-genome sequencing for children, families, and pediatric practice. Annu Rev Genomics Hum Genet 14, 535–555 (2013).
Chrystoja, C. C. & Diamandis, E. P. Whole genome sequencing as a diagnostic test: challenges and opportunities. Clin Chem 60, 724–733 (2014).
Thomas, P. D. The Gene Ontology and the Meaning of Biological Function. Methods Mol Biol 1446, 15–24 (2017).
Deng, M., Tu, Z., Sun, F. & Chen, T. Mapping Gene Ontology to proteins based on protein-protein interaction data. Bioinformatics 20, 895–902 (2004).
Al-Shahrour, F., Díaz-Uriarte, R. & Dopazo, J. Fati-GO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20, 578–580 (2004).
Lee, J. S., Katari, G. & Sachidanandam, R. GObar: a gene ontology based analysis and visualization tool for gene sets. BMC Bioinformatics 6, 189 (2005).
Bindea, G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Luthra, R., Chen, H., Roy-Chowdhuri, S. & Singh, R. R. Next-Generation Sequencing in Clinical Molecular Diagnostics of Cancer: Advantages and Challenges. Cancers 7, 2023–2036 (2015).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Giau, V.V., Bagyinszky, E., An, S.S.A. et al. Clinical genetic strategies for early onset neurodegenerative diseases. Mol. Cell. Toxicol. 14, 123–142 (2018). https://doi.org/10.1007/s13273-018-0015-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13273-018-0015-3