Abstract
Background
Euphorbia jolkini, a medicinal herb that grows on the warm beaches in Japan and South Korea, is known to be used for traditional medicines to treat a variety of ailments, including bruises, stiffness, indigestion, toothache, and diabetes.
Objective
It is to analyze the whole transcriptome and identify the genes related to the phenylpropanoid biosynthesis in the medicinally important herb E jolkini.
Methods
Paired-end Illumina HiSeq™ 2500 sequencing technology was employed for cDNA library construction and Illumina sequencing. Public databases like TAIR (The Arabidopsis Information Resource), Swissprot and KEGG (Kyoto Encyclopedia of Genes and Genomes) were used for annotations of unigenes obtained.
Results
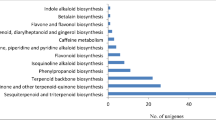
The transcriptome of E. jolkini generated 139,215 assembled transcripts with an average length of 868 bp and an N50 value of 1460 bp that were further clustered using CD-HIT into 93,801 unigenes with an average length of 847 bp (N50-1410 bp). Sixty-three percent of the coding sequences (CDS) were annotated from the longest open reading frame (ORF). A remarkable percentage of unigenes were annotated against various databases. The differentially expressed gene analysis revealed that the expression of genes related to the terpenoid backbone biosynthesis pathway was higher in the flowers, whereas that of genes related to the phenylpropanoid biosynthesis pathway was both up- and downregulated in flowers and leaves. A search of against the transcription factor domain found 1023 transcription factors (TFs) that were from 54 TF families.
Conclusion
Assembled sequences of the E. jolkini transcriptome are made available for the first time in this study E. jolkini and lay a foundation for the investigation of secondary metabolite biosynthesis.





Similar content being viewed by others
Data availability
The RNA-Seq data in this study were submitted at the NCBI public database under the accession number PRJNA627505 (https://www.ncbi.nlm.nih.gov/sra/PRJNA627505).
References
Altermann E, Klaenhammer TR (2005) PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics 6:60
Ambawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 19:307–321
Annadurai RS, Neethiraj R, Jayakumar V, Damodaran AC, Rao SN, Katta MA, Gopinathan S, Sarma SP, Senthilkumar V, Niranjan V et al (2013) De Novo transcriptome assembly (NGS) of Curcuma longa L. rhizome reveals novel transcripts related to anticancer and antimalarial terpenoids. PLoS ONE 8:e56217
Bergman ME, Davis B, Phillips MA (2019) Medically useful plant terpenoids: biosynthesis, occurrence, and mechanism of action. Molecules 24
Bhuiyan NH, Selvaraj G, Wei YD, King J (2009) Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J Exp Bot 60:509–521
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Cao D, Su YL, Yang JS (1992) Triterpene constituents from Euphorbia nematocypha Hand-Mazz. Yao xue xue bao = Acta Pharmaceutica Sinica 27:445–451
Cheng HY, Lin TC, Yang CM, Wang KC, Lin LT, Lin CC (2004) Putranjivain A from Euphorbia jolkini inhibits both virus entry and late stage replication of herpes simplex virus type 2 in vitro. J Antimicrob Chemother 53:577–583
Dagang W, Sorg B, Adolf W, Seip EH, Hecker E (1992) Oligo- and macrocyclic diterpenes in thymelaeaceae and euphorbiaceae occurring and utilized in Yunnan (Southwest China). 2. Ingenane type diterpene esters from Euphorbia nematocypha Hand.-Mazz. Phytother Res 6:237–240
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Fan H, Xiao Y, Yang Y, Xia W, Mason AS, Xia Z, Qiao F, Zhao S, Tang H (2013) RNA-Seq analysis of Cocos nucifera: transcriptome sequencing and de novo assembly for subsequent functional genomics approaches. PLoS ONE 8:e59997
Feng C, Chen M, Xu CJ, Bai L, Yin XR, Li X, Allan AC, Ferguson IB, Chen KS (2012) Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-Seq. Bmc Genomics 13:19
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–37
Garg A, Agrawal L, Misra RC, Sharma S, Ghosh S (2015) Andrographis paniculata transcriptome provides molecular insights into tissue-specific accumulation of medicinal diterpenes. BMC Genomics 16:659
Gibbons JG, Janson EM, Hittinger CT, Johnston M, Abbot P, Rokas A (2009) Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol Biol Evol 26:2731–2744
Goossens J, Mertens J, Goossens A (2016) Role and functioning of bHLH transcription factors in jasmonate signalling. J Exp Bot 68:1333–1347
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M et al (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512
Hao DC, Ge GB, Xiao PG, Zhang YY, Yang L (2011) The first insight into the tissue specific taxus transcriptome via illumina second generation sequencing. Plos One 6:e21220
He F, Pu J-X, Huang S-X, Xiao W-L, Yang L-B, Li X-N, Zhao Y, Ding J, Xu C-H, Sun H-D (2008) Eight new diterpenoids from the roots of Euphorbia nematocypha. Helv Chim Acta 91:2139–2147
Huang Y, Niu B, Gao Y, Fu L, Li W (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682
Huang CS, Luo SH, Li YL, Li CH, Hua J, Liu Y, Jing SX, Wang Y, Yang MJ, Li SH (2014) Antifeedant and antiviral diterpenoids from the fresh roots of Euphorbia jolkinii. Nat Prod Bioprospect 4:91–100
Inha Y (2011) 제주의 5월 암대극과 검은딱새. https://www.leisuretimes.co.kr/news/articleView.html?idxno=22288. Accessed 10 Dec 2018
Jassbi AR (2006) Chemistry and biological activity of secondary metabolites in Euphorbia from Iran. Phytochemistry 67:1977–1984
Kanehisa M (2019) Toward understanding the origin and evolution of cellular organisms. Protein Sci 28:1947–1951
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kim J-Y, Lee J-A, Yoon W-J, Oh D-J, Jung Y-H, Lee W-J, Park S-Y (2006a) Antioxidative and antimicrobial activities of Euphorbia jolkini extracts. Korean J Food Sci Technol 38:699–706
Kim J-Y, Lee J-A, Yoon W-J, Oh D-J, Jung Y-H, Lee W-J, Park S-Y (2006b) Antioxidative and antimicrobial activities of Euphorbia jolkinii extracts. Korean J Food Sci Technol 38:699–706
Kim M-J, Seoyeon K, Hyun KH, Kim DS, Kim S-Y, Hyun C-G (2017) Effects of Rumex axetosella, Sonchus oleraceus and Euphoibia jolkini extracts on melanin synthesis in melanoma cells. Korean Soc Biotechnol Bioeng J 32:187–192
Kim J-A, Roy NS, Lee IH, Choi AY, Choi BS, Yu YS, Park NI, Park KC, Kim S, Yang HS et al (2018) Genome-wide transcriptome profiling of the medicinal plant Zanthoxylum planispinum using a single-molecule direct RNA sequencing approach. Genomics 111:973–979
Koutaniemi S, Warinowski T, Karkonen A, Alatalo E, Fossdal CG, Saranpaa P, Laakso T, Fagerstedt KV, Simola LK, Paulin L et al (2007) Expression profiling of the lignin biosynthetic pathway in Norway spruce using EST sequencing and real-time RT-PCR. Plant Mol Biol 65:311–328
Kuo PL, Cho CY, Hsu YL, Lin TC, Lin CC (2006) Putranjivain A from Euphorbia jolkini inhibits proliferation of human breast adenocarcinoma MCF-7 cells via blocking cell cycle progression and inducing apoptosis. Toxicol Appl Pharmacol 213:37–45
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J (2001) Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57:1187–1195
Lee SH, Tanaka T, Nonaka G, Nishioka I (2004) Structure and biogenesis of jolkinin, a highly oxygenated ellagitannin from Euphorbia jolkinii. J Nat Prod 67:1018–1022
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323
Lulin H, Xiao Y, Pei S, Wen T, Shangqin H (2012) The first Illumina-based de novo transcriptome sequencing and analysis of safflower flowers. PLoS ONE 7:e38653
Maru N, Chikaraishi N, Yokota K, Kawazoe Y, Uemura D (2013) Jolkinolide F, a cytotoxic diterpenoid from Euphorbia jolkinii. Chem Lett 42:756–757
Menden B, Kohlhoff M, Moerschbacher BM (2007) Wheat cells accumulate a syringyl-rich lignin during the hypersensitive resistance response. Phytochemistry 68:513–520
Moerschbacher BM, Noll U, Gorrichon L, Reisener HJ (1990) Specific inhibition of lignification breaks hypersensitive resistance of wheat to stem rust. Plant Physiol 93:465–470
Naoumkina MA, Zhao Q, Gallego-Giraldo L, Dai X, Zhao PX, Dixon RA (2010) Genome-wide analysis of phenylpropanoid defence pathways. Mol Plant Pathol 11:829–846
Park SB, Kim MS, Lee HS, Lee SH, Kim SH (2010) 1,2,3,6-tetra-O-galloyl-beta-D-allopyranose gallotannin isolated, from Euphorbia jolkini, attenuates LPS-induced nitric oxide production in macrophages. Phytother Res 24:1329–1333
Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25
Roy NS, Kim JA, Choi AY, Ban YW, Park NI, Park KC, Yang HS, Choi IY, Kim S (2018) RNA-seq de novo assembly and differential transcriptome analysis of Korean medicinal herb Cirsium japonicum var spinossimum. Genomics Inform 16:e34
Soltani Howyzeh M, Sadat Noori SA, Shariati JV, Amiripour M (2018) Comparative transcriptome analysis to identify putative genes involved in thymol biosynthesis pathway in medicinal plant Trachyspermum ammi L. Sci Rep 8:13405
Tholl D (2015) Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol 148:63–106
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wang C, Peng D, Zhu J, Zhao D, Shi Y, Zhang S, Ma K, Wu J, Huang L (2019) Transcriptome analysis of Polygonatum cyrtonema Hua: identification of genes involved in polysaccharide biosynthesis. Plant Methods 15:65
Xie Z, Nolan TM, Jiang H, Yin Y (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10:228
Yang Y, Xu M, Luo Q, Wang J, Li H (2014) De novo transcriptome analysis of Liriodendron chinense petals and leaves by Illumina sequencing. Gene 534:155–162
Yuan X, Wang H, Cai J, Li D, Song F (2019) NAC transcription factors in plant immunity. Phytopathol Res 1:3
Zhang L, Jia H, Yin Y, Wu G, Xia H, Wang X, Fu C, Li M, Wu J (2013) Transcriptome analysis of leaf tissue of Raphanus sativus by RNA sequencing. PLoS ONE 8:e80350
Acknowledgements
This work was supported by the National Institute of Biological resources, Ministry of Environment (MOE) of the Republic of Korea (NIBR201830101, NIBR201921101).
Author information
Authors and Affiliations
Contributions
SK and IYC conceived and designed the project. JK, SK and NIP collected plants and prepared sample materials for genome analysis. IL, JK, JHY, NIP, KCP and NSR performed experiment. RVR, NSR analysed the data. NSR was a major contributor to write the manuscript. SK, KCP and IYC corrected and completed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest in regards to the content of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Roy, N.S., Lee, Ih., Kim, JA. et al. De novo assembly and characterization of transcriptome in the medicinal plant Euphorbia jolkini. Genes Genom 42, 1011–1021 (2020). https://doi.org/10.1007/s13258-020-00957-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-020-00957-1