Abstract
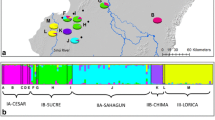
To optimize conservation efforts, it is necessary to determine the risk of extinction by collecting reliable population information for a given species. We developed eight novel, polymorphic microsatellite markers and used these markers in conjunction with twelve existing markers to measure genetic diversity of South Korean populations of leopard cat (Prionailurus bengalensis), a species for which population size and habitat area data are unknown in the country, to assess its conservation status. The average number of alleles and the observed heterozygosity of the species were 3.8 and 0.41, respectively, and microsatellite diversity was lower than the average genetic diversity of 57 populations of 12 other felid species, and lower than that of other mammal populations occurring in South Korea, including the raccoon dog (Nyctereutes procyonoides), water deer (Hydropotes inermis), and endangered long-tailed goral (Naemorhedus caudatus). Furthermore, analysis of genetic structure in the national leopard cat population showed no clear genetic differentiation, suggesting that it is not necessary to divide the South Korean leopard cat population into multiple management units for the purposes of conservation. These results indicate that the genetic diversity of the leopard cat in South Korea is unexpectedly low, and that the risk of local extinction is, as a result, substantial. Thus, it is necessary to begin appropriate conservation efforts at a national level to conserve the leopard cat population in South Korea.



Similar content being viewed by others
References
Anderson Jr CR, Lindzey FG, McDonald DB (2004) Genetic structure of cougar populations across the Wyoming Basin: metapopulation or megapopulation. J Mammal 85:1207–1214
Antunes A, Troyer JL, Roelke ME, Pecon-Slattery J, Packer C, Winterbach C, Winterbach H, Hemson G, Frank L, Stander P (2008) The evolutionary dynamics of the lion Panthera leo revealed by host and viral population genomics. PLoS genetics 4:e1000251
Baillie J, Hilton-Taylor C, Stuart SN (2004) 2004 IUCN red list of threatened species: a global species assessment. IUCN, Gland
Bang S, Ahn S (2005) Development of red list categories and criteria for the protection of Endangered Species in Korea. Korea Environment Institute, Seoul
Buckley-Beason VA, Johnson WE, Nash WG, Stanyon R, Menninger JC, Driscoll CA, Howard J, Bush M, Page JE, Roelke ME (2006) Molecular evidence for species-level distinctions in clouded leopards. Current Biology 16:2371–2376
Ceballos G, Ehrlich PR (2002) Mammal population losses and the extinction crisis. Science 296:904–907
Chang C, Lee H, Park T, Kim H (2005) Reconsideration of rare and endangered plant species in Korea based on the IUCN Red List categories. Korean J Ecol 28:305–320
Chen C, Durand E, Forbes F, François O (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes 7:747–756
Choi T (2007) Road-kill mitigation strategies for mammals in Korea: data based on surveys of road-kill, non-wildlife passage use, and home-range. PhD Dissertation, Seoul National University
Choi T, Kwon H, Woo D, Park C (2012) Habitat selection and management of the leopard cat (Prionailurus bengalensis) in a rural area of Korea. Korean J Environ Ecol 26:322–332
Choi SK, Chun S, An J, Lee M, Kim HJ, Min M, Kwon S, Choi TY, Lee H, Kim KS (2015) Genetic diversity and population structure of the long-tailed goral, Naemorhedus caudatus, in South Korea. Genes Genet Syst 90:31–41
Dalton DL, Charruau P, Boast L, Kotzé A (2013) Social and genetic population structure of free-ranging cheetah in Botswana: implications for conservation. Eur J Wildl Res 59:281–285
Doyle JM, Hacking CC, Willoughby JR, Sundaram M, DeWoody JA (2015) Mammalian genetic diversity as a function of habitat, body size, trophic class, and conservation status. J Mammal 96:564–572
Dutta T, Sharma S, Maldonado JE, Wood TC, Panwar H, Seidensticker J (2013) Fine-scale population genetic structure in a wide-ranging carnivore, the leopard (Panthera pardus fusca) in central India. Divers Distrib 19:760–771
Eo SH, Ko BJ, Lee B, Seomun H, Kim S, Kim M, Kim JH, An J (2016a) A set of microsatellite markers for population genetics of leopard cat (Prionailurus bengalensis) and cross-species amplification in other felids. Biochem Syst Ecol 66:196–200
Eo SH, Lee W, Lee B, Ko BJ, Kim JU, Jeon JH (2016b) Microsatellite markers for the Ussuri white-toothed shrew (Soricidae: Crocidura lasiura) developed by Ion Torrent sequencing and their application to the shrew populations in disturbed forests. Genes Genom 38:351–357
Epps CW, Palsbøll PJ, Wehausen JD, Roderick GK, Ramey RR, McCullough DR (2005) Highways block gene flow and cause a rapid decline in genetic diversity of desert bighorn sheep. Ecol Lett 8:1029–1038
Evans SR, Sheldon BC (2008) Interspecific patterns of genetic diversity in birds: correlations with extinction risk. Conserv Biol 22:1016–1025
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Faircloth BC (2008) Msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour 8:92–94
Faircloth BC, Reid A, Valentine T, Eo SH, Terhune TM, Glenn TC, Palmer WE, Nairn CJ, Carroll JP (2005) Tetranucleotide, trinucleotide, and dinucleotide loci from the bobcat (Lynx rufus). Mol Ecol Resour 5:387–389
Frankham R (2012) How closely does genetic diversity in finite populations conform to predictions of neutral theory? Large deficits in regions of low recombination. Heredity 108:167–178
Frankham R, Ballou JD, Briscoe DA (2004) A primer of conservation genetics. Cambridge University Press, Cambridge
Frankham R, Bradshaw CJ, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Garner A, Rachlow JL, Hicks JF (2005) Patterns of genetic diversity and its loss in mammalian populations. Conserv Biol 19:1215–1221
Gaur A, Shailaja K, Singh A, Arunabala V, Satyarebala B, Singh L (2006) Twenty polymorphic microsatellite markers in the Asiatic lion (Panthera leo persica). Conserv Genet 7:1005–1008
Gottelli D, Wang J, Bashir S, Durant SM (2007) Genetic analysis reveals promiscuity among female cheetahs. Proc Biol Sci 274:1993–2001
Haag T, Santos A, Sana D, Morato R, Cullen Jr L, Crawshaw Jr P, De Angelo C, Di Bitetti M, Salzano F, Eizirik E (2010) The effect of habitat fragmentation on the genetic structure of a top predator: loss of diversity and high differentiation among remnant populations of Atlantic Forest jaguars (Panthera onca). Mol Ecol 19:4906–4921
Hellborg L, Walker CW, Rueness EK, Stacy JE, Kojola I, Valdmann H, Vilà C, Zimmermann B, Jakobsen KS, Ellegren H (2002) Differentiation and levels of genetic variation in northern European lynx (Lynx lynx) populations revealed by microsatellites and mitochondrial DNA analysis. Conserv Genet 3:97–111
Holderegger R, Di Giulio M (2010) The genetic effects of roads: a review of empirical evidence. Basic Appl Ecol 11:522–531
Hong Y, Kim K, Lee H, Min M (2013) Population genetic study of the raccoon dog (Nyctereutes procyonoides) in South Korea using newly developed 12 microsatellite markers. Genes Genet Syst 88:69–76
Kim H, Lee BC, Kim YS, Chang C (2012) Critiques of the endangered and protected wild species list in Korea’ proposed by Korea Ministry of Environment and listing process-Is this the best process for the current national management of endangered wildlife and plants in Korea?- J Korean For Soc 101:7–19
Lee BH, Won CM, Cho YS, Han SH (2008) Taxonomy status and geographic variations of Prionailurus Bengalenesis in Korea. National Institute of Biological Resources, Incheon, Incheon
Lee YS, Choi SK, An J, Park HC, Kim SI, Min MS, Kim KS, Lee H (2011) Isolation and characterization of 12 microsatellite loci from Korean water deer (Hydropotes inermis argyropus) for population structure analysis in South Korea. Genes Genom 33:535–540
Mattucci F, Oliveira R, Bizzarri L, Vercillo F, Anile S, Ragni B, Lapini L, Sforzi A, Alves P, Lyons L, Randi E (2013) Genetic structure of wildcat (Felis silvestris) populations in Italy. Ecol Evol 3:2443–2458
Moritz C (1994) Defining ‘evolutionarily significant units’ for conservation. Trends Ecol Evol 9:373–375
Park S, Kim H, Chang C (2013) Assessing red list categories to a Korean endangered species based on IUCN criteria-Hanabusaya asiatica (Nakai) Nakai. Korean J Plant Taxon 43:128–138
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28:2537–2539
Pinsky ML, Palumbi SR (2014) Meta-analysis reveals lower genetic diversity in overfished populations. Mol Ecol 23:29–39
Rivers MC, Brummitt NA, Lughadha EN, Meagher TR (2014) Do species conservation assessments capture genetic diversity? Glob Ecol Conserv 2:81–87
Rodrigues AS, Pilgrim JD, Lamoreux JF, Hoffmann M, Brooks TM (2006) The value of the IUCN Red List for conservation. Trends Ecol Evol 21:71–76
Rodzen JA, Banks J, Meredith E, Jones K (2007) Characterization of 37 microsatellite loci in mountain lions (Puma concolor) for use in forensic and population applications. Conserv Genet 8:1239–1241
Ross J, Brodie J, Cheyne S, Hearn A, Izawa M, Loken B, Lynam A, McCarthy J, Mukherjee S, Phan C, Rasphone A, Wilting A (2015) Prionailurus bengalensis. The IUCN Red List of Threatened Species 2015: e. T18146A50661611
Rousset F (2008) Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa
Rueness EK, Stenseth NC, O'donoghue M, Boutin S, Ellegren H, Jakobsen KS (2003) Ecological and genetic spatial structuring in the Canadian lynx. Nature 425:69–72
Schmidt K, Kowalczyk R, Ozolins J, Männil P, Fickel J (2009) Genetic structure of the Eurasian lynx population in north-eastern Poland and the Baltic states. Conserv Genet 10:497–501
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Sharma R, Stuckas H, Moll K, Khan I, Bhaskar R, Goyal S, Tiedemann R (2008) Fourteen new di-and tetranucleotide microsatellite loci for the critically endangered Indian tiger (Panthera tigris tigris). Mol Ecol Resour 8:1480–1482
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Spong G, Johansson M, Björklund M (2000) High genetic variation in leopards indicates large and long-term stable effective population size. Mol Ecol 9:1773–1782
Steffens DL, Sutter SL, Roemer SC (1993) An alternate universal forward primer for improved automated DNA sequencing of M13. BioTechniques 15:580–582
Traill LW, Bradshaw CJ, Brook BW (2007) Minimum viable population size: a meta-analysis of 30 years of published estimates. Biol Conserv 139:159–166
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Waits LP, Buckley-Beason VA, Johnson WE, Onorato D, McCarthy T (2007) A select panel of polymorphic microsatellite loci for individual identification of snow leopards (Panthera uncia). Mol Ecol Resour 7:311–314
Wiseman R, O’Ryan C, Harley E (2000) Microsatellite analysis reveals that domestic cat (Felis catus) and southern African wild cat (F. lybica) are genetically distinct. Anim Conserv Forum 3:221–228
Won C, Smith KG (1999) History and current status of mammals of the Korean Peninsula. Mamm Rev 29:3–36
Wu J, Lei Y, Fang S, Wan Q (2009) Twenty-one novel tri-and tetranucleotide microsatellite loci for the Amur tiger (Panthera tigris altaica). Conserv Genet 10:567–570
Yashima AS, Innan H (2016) VarVer: a database of microsatellite variation in vertebrates. Mol Ecol Resour 17:824–833
Youngquist MB, Inoue K, Berg DJ, Boone MD (2017) Effects of land use on population presence and genetic structure of an amphibian in an agricultural landscape. Landsc Ecol 32:147–162
Zhang Z, Zhang W, Yue B, Shen F, Zhang L, Hou R, Zhu M (2006) Twelve polymorphic microsatellite loci for the South China tiger Panthera tigris amoyensis. Mol Ecol Resour 6:24–26
Acknowledgements
We thank members of the Eo lab for their comments on the manuscript. This work was supported by a grant from the Kongju National University in 2013 and a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR201603103).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Byung June Ko, Junghwa An, Hong Seomun, Mu-Yeong Lee, and Soo Hyung Eo declares that they have no conflict of interest.
Ethical approval
All animal experiments throughout the study were conducted in accordance with guidelines of The Ministry of Environment for the care and use of animals.
Rights and permissions
About this article
Cite this article
Ko, B.J., An, J., Seomun, H. et al. Microsatellite DNA analysis reveals lower than expected genetic diversity in the threatened leopard cat (Prionailurus bengalensis) in South Korea. Genes Genom 40, 521–530 (2018). https://doi.org/10.1007/s13258-018-0654-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-018-0654-8