Abstract
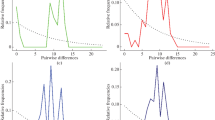
Domestic goats (Capra hircus) are classified into six different mitochondrial DNA (mtDNA) haplogroups (A, B, C, D, F, and G), of which haplogroup A shows worldwide distribution. However, there is currently no key for genetic and geographic classification of haplogroup A. In this study, we determined the complete sequences of the mtDNA control region from Korean native goat breeds. In addition, we compared these sequences with those of other domestic goat breeds/populations globally. A distinct variation site was found at position 717. Of two types (C and T) from this variation, C type sequences were found only in haplogroup A, and were specific to Eastern Asia. On the other hand, T type sequences were distributed in all haplogroups. Phylogenetic analysis divided the two types according to different phylogenetic locations. These results suggest that a variation in position 717 is a key for geographical classification of mtDNA haplogroup A. Moreover, these distribution patterns might provide for identification of the maternal origin of goat genetic resources, including Korean native goat breeds.




Similar content being viewed by others
References
An JH, Okumura H, Lee YS, Kim KS, Min MS, Lee H (2010) Organization and variation of the mitochondrial DNA control region in five Caprinae species. Genes Genom 32:335–344
Chen SY, Su YH, Wu SF, Sha T, Zhang YP (2005) Mitochondrial diversity and phylogeographic structure of Chinese domestic goats. Mol Phylogenet Evol 37:804–814
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hassanin A, Bonillo C, Nquyen BX, Cruaud C (2010) Comparisons between mitochondrial genomes of domestic goat (Capra hircus) reveal the presence of numts and multiple sequencing errors. Mitochondria DNA 21:68–76
Joshi MB, Rout PK, Mandal AK, Tyler-Smith C, Singh L, Thangaraj K (2004) Phylogeography and origin of Indian domestic goats. Mol Biol Evol 21:454–462
Kim JH, Cho CY, Choi SB, Cho YM, Yeon SH, Yang BS (2011) mtDNA diversity and phylogenetic analysis of Korean native goats. Korean J Life Sci 21:1329–1335
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lin BZ, Odahara S, Ishida M, Kato T, Sasazaki S, Nozawa K, Mannen H (2013) Molecular phylogeography and genetic diversity of East Asian goats. Anim Genet 44:79–85
Liu YP, Cao SX, Chen SY, Yao YG, Liu TZ (2009) Genetic diversity of Chinese domestic goat based on the mitochondrial DNA sequence variation. J Anim Breed Genet 126:80–89
Luikar G, Gielly L, Excoffier L, Vigne JD, Bouvet J, Taberlet P (2001) Multiple maternal origins and weak phylogeographic structure in domestic goats. Proc Natl Acad Sci USA 98:5927–5932
Mahmoudi B, Panahi B, Mohammadi SA, Daliri M, Babayev MSh (2014) Microsatellite based phylogeny and bottleneck studies of Iranian indigenous goat populations. Anim Biotechnol 25:210–222
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Naderi S, Rezaei HR, Taberlet P, Zundel S, Rafat SA, Naghash HR, El-Barody MAA, Ertugrul O, Pompanon F (2007) Large-scale mitochondrial DNA analysis of the domestic goat reveals six haplogroups with high diversity. PLoS One 10:e1012
Odahara S, Chung HJ, Choi SH, Yu SL, Sasazaki S, Mannen H, Park CS, Lee JH (2005) Mitochondrial DNA diversity of Korean native goats. J Anim Sci 19:482–485
Pereira F, Queirós S, Gusmão L, Nijman I, Cuppen E, Lenstra JA, Econogene Consortium, Davis SJ, Nejmeddine F, Amorim A (2009) Tracing the history of goat pastoralism: new clues from mitochondrial and Y chromosome DNA in North Africa. Mol Biol Evol 26:2765–2773
Porter V (1996) Goats of the world. Farming Press, Ipswich
Porter V, Tebbit J (1996) Goats of the World. Farming Press Limited, Ipswich
Pringle H (1998) Neolithic agriculture: reading the signs of ancient animal domestication. Science 282:1448
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406
Sardina MT, Ballester M, Marmi J, Finocchiaro R, van Kaam JB (2006) Phylogenetic analysis of Sicilian goats reveals a new mtDNA lineage. Anim Genet 34:376–378
Sultana S, Mannen H, Tsuji S (2003) Mitochondrial DNA diversity of Pakistani goats. Anim Genet 34:417–421
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetic analysis using maximum likelihood evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Higgins DG, Gibson TI (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Tu YR, Jiang Y, Han ZY, Feng WQ (1989) Sheep and goat breeds in China. Shanghai Scientific and Technical Publishers, Shanghai, pp 1–25
Zeder MA, Hesse B (2000) The initial domestication of goats (Capra hircus) in the Zagros mountain 10,000 years ago. Science 287:2254–2257
Zhao YZ (1995) Goat production. Chinese Agricultural Press, Beijing, pp 24–26
Zhong T, Zhao QJ, Niu LL, Wang J, Jin PF, Zhao W, Wang LJ, Li L, Zhang HP, Ma YH (2013) Genetic phylogeography and maternal lineages of 18 Chinese black goat breeds. Trop Anim Health Prod 45:1833–1837
Acknowledgments
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ00843101)” Rural Development Administration, Republic of Korea.
Conflict of interest
The authors have declared no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, JH., Byun, MJ., Choi, SB. et al. Detection of a distinct variation site for geographical classification of mitochondrial DNA haplogroup A in the domestic goat (Capra hircus). Genes Genom 36, 701–709 (2014). https://doi.org/10.1007/s13258-014-0204-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-014-0204-y