Abstract
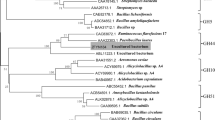
Sequence diversity within a family of functional enzymes provides a platform for novel gene development and protein engineering to improve the properties of these enzymes for further applications. Glycoside hydrolase family 48 (GH48) is an important group of microbial cellulases. However, the genetic diversity and gene discovery of GH48 enzyme in natural environments are rarely reported. In this study, the genetic diversity of GH48 from Changbai Mountain soil was evaluated by building a clone library via a culture-independent molecular method for the first time. Results showed that the genetic diversity of GH48 in Changbai Mountain soil was different from that in thermophilic compost and marine sediment libraries, and more than 80% of the sequences exhibited the highest identity with cellulase genes from Chloroflexi. Novel GH48 genes were also cloned, and the recombinants Cel48_hm01 and Cel48_hm02 were prokaryotically expressed, purified, and characterized. Characterization results suggested that they were probably endocellulases that adopted a catalytic mechanism similar to the GH48 cellulase from Clostridium. This study revealed the genetic distribution of glycoside hydrolases in soil environment, described Changbai Mountain soil as a valuable source for glycoside hydrolase gene screening, and presented supplementary property data on novel GH48 from natural soil environments.





Similar content being viewed by others
References
Ahmad A, Niwa Y, Goto S, Kobayashi K, Shimizu M, Ito S, Usui Y, Nakayama T, Kobayashi H (2015) Genome-wide screening of salt tolerant genes by activation-tagging using dedifferentiated calli of Arabidopsis and its application to finding gene for Myo-inositol-1-P-synthase. PLoS One 10:e0115502
Barbi F, Bragalini C, Vallon L, Prudent E, Dubost A, Fraissinet-Tachet L, Marmeisse R, Luis P (2014) PCR primers to study the diversity of expressed fungal genes encoding lignocellulolytic enzymes in soils using high-throughput sequencing. PLoS One 9:e116264
Berger E, Zhang D, Zverlov VV, Schwarz WH (2007) Two noncellulosomal cellulases of Clostridium thermocellum, Cel9I and Cel48Y, hydrolyse crystalline cellulose synergistically. FEMS Microbiol Lett 268:194–201
Berlemont R, Martiny AC (2013) Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol 79:1545–1554
Berlemont R, Martiny AC (2015) Genomic potential for polysaccharide deconstruction in bacteria. Appl Environ Microbiol 81:1513–1519
Book AJ, Lewin GR, McDonald BR, Takasuka TE, Doering DT, Adams AS, Blodgett JAV, Clardy J, Raffa KF, Fox BG, Currie CR (2014) Cellulolytic Streptomyces strains associated with herbivorous insects share a phylogenetically linked capacity to degrade lignocellulose. Appl Environ Microbiol 80:4692–4701
Borrelli GM, Trono D (2015) Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int J Mol Sci 16:20774–20840
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
De Menezes AB, Prendergast-Miller MT, Poonpatana P, Farrell M, Bissett A, Macdonald LM, Toscas P, Richardson AE, Thrall PH (2015) C/N ratio drives soil actinobacterial cellobiohydrolase gene diversity. Appl Environ Microbiol 81:3016–3028
Devillard E, Goodheart DB, Karnati SKR, Bayer EA, Lamed R, Miron J, Nelson KE, Morrison M (2004) Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture. J Bacteriol 186:136–145
Guimarães BG, Souchon H, Lytle BL, David Wu JH, Alzari PM (2002) The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum cellulosome. J Mol Biol 320:587–596
Hamid SBA, Islam MM, Das R (2015) Cellulase biocatalysis: key influencing factors and mode of action. Cellulose 22:2157–2182
Hua M, Zhao SB, Zhang LL, Liu DB, Xia HM, Li F, Chen S (2015) Direct detection, cloning and characterization of a glucoside hydrolase from forest soil. Biotechnol Lett 37:1227–1232
Huang H, Wang G, Zhao Y, Shi P, Luo H, Yao B (2010) Direct and efficient cloning of full-length genes from environmental DNA by RT-qPCR and modified TAIL-PCR. Appl Microbiol Biotechnol 87:1141–1149
Irwin DC, Zhang S, Wilson DB (2000) Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur J Biochem 267:4988–4997
Izquierdo JA, Sizova MV, Lynd LR (2010) Diversity of bacteria and glycosyl hydrolase family 48 genes in cellulolytic consortia enriched from thermophilic biocompost. Appl Environ Microbiol 76:3545–3553
Jain L, Agrawal D (2018) Performance evaluation of fungal cellulases with dilute acid pretreated sugarcane bagasse: a robust bioprospecting strategy for biofuel enzymes. Renew Energy 115:978–988
Ji S, Wang S, Tan Y, Chen X, Schwarz W, Li F (2012) An untapped bacterial cellulolytic community enriched from coastal marine sediment under anaerobic and thermophilic conditions. FEMS Microbiol Lett 335:39–46
Kiss H, Nett M, Domin N, Martin K, Maresca JA, Copeland A, Lapidus A, Lucas S, Berry KW, Glavina Del Rio T, Dalin E, Tice H, Pitluck S, Richardson P, Bruce D, Goodwin L, Han C, Detter JC, Schmutz J, Brettin T, Land M, Hauser L, Kyrpides NC, Ivanova N, Göker M, Woyke T, Klenk HP, Bryant DA (2011) Complete genome sequence of the filamentous gliding predatory bacterium Herpetosiphon aurantiacus type strain (114-95T). Stand Genomic Sci 5:356–370
Kostylev M, Alahuhta M, Chen M, Brunecky R, Himmel ME, Lunin VV, Brady J, Wilson DB (2014) Cel48A from Thermobifida fusca: structure and site directed mutagenesis of key residues. Biotechnol Bioeng 111:664–673
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kyeremeh IA, Charles CJ, Rout SP, Laws AP, Humphreys PN (2016) Microbial community evolution is significantly impacted by the use of calcium isosaccharinic acid as an analogue for the products of alkaline cellulose degradation. PLoS One 11:e0165832
Li L, Xing M, Lv J, Wang X, Chen X (2017) Response of rhizosphere soil microbial to Deyeuxia angustifolia encroaching in two different vegetation communities in alpine tundra. Sci Rep 7:43150
Liu YG, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Olson DG, Tripathi SA, Giannone RJ, Lo J, Caiazza NC, Hogsett DA, Hettich RL, Guss AM, Dubrovsky G, Lynd LR (2010) Deletion of the Cel48S cellulase from Clostridium thermocellum. Proc Natl Acad Sci 107:17727–17732
Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, Ståhlberg J, Beckham GT (2015) Fungal cellulases. Chem Rev 115:1308–1448
Pereyra LP, Hiibel SR, Prieto Riquelme MV, Reardon KF, Pruden A (2010) Detection and quantification of functional genes of cellulose-degrading, fermentative, and sulfate-reducing bacteria and methanogenic archaea. Appl Environ Microbiol 76:2192–2102
Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ Jr, Hallett JP, Leak DJ, Liotta CL, Mielenz JR, Murphy R, Templer R, Tschaplinski T (2006) The path forward for biofuels and biomaterials. Science 311:484–489
Reverbel-Leroy C, Pages S, Belaich A, Belaich JP, Tardif C (1997) The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J Bacteriol 179:46–52
Santiago M, Ramírez-Sarmiento CA, Zamora RA, Parra LP (2016) Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front Microbiol 7:140
Saraihom S, Kobayashi DY, Lotrakul P, Prasongsuk S, Eveleigh DE, Punnapayak H (2016) First report of a tropical Lysobacter enzymogenes producing bifunctional endoglucanase activity towards carboxymethylcellulose and chitosan. Ann Microbiol 66:907–919
Schumann P, Cui X, Stackebrandt E, Kroppenstedt RM, Xu L, Jiang C (2004) Jonesia quinghaiensis sp. nov., a new member of the suborder Micrococcineae. Int J Syst Evol Microbiol 54:2181–2184
Segato F, Damásio ARL, De Lucas RC, Squina FM, Prade RA (2014) Genomics review of holocellulose deconstruction by aspergilli. Microbio Mol Biol Rev 78:588–613
Shen C, Xiong J, Zhang H, Feng Y, Lin X et al (2013) Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem 57:204–211
Sheng P, Li Y, Marshall SDG, Zhang H (2015) High genetic diversity of microbial cellulase and hemicellulase genes in the hindgut of Holotrichia parallela larvae. Int J Mol Sci 16:16545–16559
Sizova MV, Izquierdo JA, Panikov NS, Lynd LR (2011) Cellulose-and xylan-degrading thermophilic anaerobic bacteria from biocompost. Appl Environ Microbiol 77:2282–2291
Steenbakkers PJ, Freelove A, Van Cranenbroek B, Sweegers BM, Harhangi HR, Vogels GD, Hazlewood GP, Gilbert HJ, Op Den Camp HJM (2002) The major component of the cellulosomes of anaerobic fungi from the genus Piromyces is a family 48 glycoside hydrolase. DNA Seq 13:313–320
Still PC, Johnson TA, Theodore CM, Loveridge ST, Crews P (2014) Scrutinizing the scaffolds of marine biosynthetics from different source organisms: Gram-negative cultured bacterial products enter center stage. J Nat Prod 77:690–602
Sukharnikov LO, Alahuhta M, Brunecky R, Upadhyay A, Himmel ME, Lunin VV, Zhulin IB (2012) Sequence, structure, and evolution of cellulases in glycoside hydrolase family 48. J Biol Chem 287:41068–41077
Takasaki K, Miura T, Kanno M, Tamaki H, Hanada S, Kamagata Y, Kimura N (2013) Discovery of glycoside hydrolase enzymes in an Avicel adapted forest soil fungal community by a metatranscriptomic approach. PLoS One 8:e55485
Talamantes D, Biabini N, Dang H, Abdoun K, Berlemont R (2016) Natural diversity of cellulases, xylanases, and chitinases in bacteria. Biotechnol Biofuels 9:133
Wang F, Xu XX, Qu Z, Wang C, Lin HP, Xie QY, Ruan JS, Sun M, Hong K (2011) Nonomuraea wenchangensis sp. nov., isolated from mangrove rhizosphere soil. Int J Syst Evol Micrbiol 61:1304–1308
Wang G, Huang X, Ng TB, Lin J, Ye XY (2014) High phylogenetic diversity of glycosyl hydrolase family 10 and 11 xylanases in the sediment of Lake Dabusu in China. PLoS One 9:e112798
Wang GZ, Wang YR, Yang PL, Luo HY, Huang HQ, Shi PJ, Meng K, Yao B (2010) Molecular detection and diversity of xylanase genes in alpine tundra soil. Appl Microbiol Biotechnol 87:1383–1393
Zhang Q, Liang G, Guo T, He P, Wang X, Zhou W (2017) Evident variations of fungal and actinobacterial cellulolytic communities associated with different humified particle-size fractions in a long-term fertilizer experiment. Soil Biol Biochem 113:1–13
Zhao W, Zhong Y, Yuan H, Wang J, Zheng H, Wang Y, Cen X, Xu F, Bai J, Han X, Lu G, Zhu Y, Shao Z, Yan H, Li C, Peng N, Zhang Z, Zhang Y, Lin W, Fan Y, Qin Z, Hu Y, Zhu B, Wang S, Ding X, Zhao GP (2010) Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res 20:1096–1008
Zhou JP, Huang HQ, Meng K, Shi PJ, Wang Y, Luo HY, Yang PL, Bai YG, Yao B (2010) Cloning of a new xylanase gene from Streptomyces sp. TN119 using a modified thermal asymmetric interlaced-PCR specific for GC-rich genes and biochemical characterization. Appl Biochem Biotechnol 160:1277–1292
Zou Y, Sang W, Wang S, Thomas EW, Liu Y, Yu Z, Wang C, Axmacher JC (2015) Diversity patterns of ground beetles and understory vegetation in mature, secondary, and plantation forest regions of temperate Northern China. Ecol Evol 5:531–542
Acknowledgments
This work was supported by the National Natural Science Foundation of Jilin, China (No. 20140101137JC).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Hua, M., Yu, S., Ma, Y. et al. Genetic diversity detection and gene discovery of novel glycoside hydrolase family 48 from soil environmental genomic DNA. Ann Microbiol 68, 163–174 (2018). https://doi.org/10.1007/s13213-018-1327-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-018-1327-1