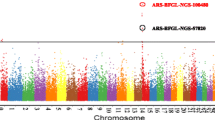
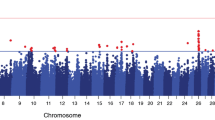
Abstract
This study elucidated potential genetic variants and QTLs associated with clinical mastitis incidence traits in Bos indicus breed, Sahiwal. Estimated breeding values for the traits (calculated using Bayesian inference) were used as pseudo-phenotypes for association with genome-wide SNPs and further QTL regions underlying the traits were identified. In all, 25 SNPs were found to be associated with the traits at the genome-wide suggestive threshold (p ≤ 5 × 10−4) and these SNPs were used to define QTL boundaries based on the linkage disequilibrium structure. A total of 16 QTLs were associated with the trait EBVs including seven each for clinical mastitis incidence (CMI) in first and second lactations and two for CMI in third lactation. Nine out of sixteen QTLs overlapped with the already reported QTLs for mastitis traits, whereas seven were adjudged as novel ones. Important candidates for clinical mastitis in the identified QTL regions included DNAJB9, ELMO1, ARHGAP26, NR3C1, CACNB2, RAB4A, GRB2, NUP85, SUMO2, RBPJ, and RAB33B genes. These findings shed light on the genetic architecture of the disease in Bos indicus, and present potential regions for fine mapping and downstream analysis in future.

Similar content being viewed by others
Data availability
The phenotypic as well as genotypic data used in the present study are available with the https://doi.org/10.6084/m9.figshare.16570587 and would be made accessible based on individual requests to the corresponding author after due consideration.
References
Adorisio S, Fierabracci A, Muscari I, Liberati AM, Ayroldi E, Migliorati G, Thuy TT, Riccardi C, Delfino DV (2017) SUMO proteins: guardians of immune system. J Autoimmun 84:21–28. https://doi.org/10.1016/j.jaut.2017.09.001
Ashwell MS, Heyen DW, Weller JI, Ron M, Sonstegard TS, Van Tassell CP, Lewin HA (2005) Detection of quantitative trait loci influencing conformation traits and calving ease in Holstein–Friesian cattle. J Dairy Sci 88:4111–4119. https://doi.org/10.3168/jds.S0022-0302(05)73095-2
Boichard D, Boussaha M, Capitan A, Rocha D, Hozé C, Sanchez MP, Tribout T, Letaief R, Croiseau P, Grohs C et al (2018) Experience from large scale use of the EuroGenomics custom SNP chip in cattle. Proc World Congr Genet Appl Livest Prod 11:675
Burton JL, Erskine RJ (2003) Immunity and mastitis. Some new ideas for an old disease. Vet Clin N Am Food Anim Pract 19:1. https://doi.org/10.1016/s0749-0720(02)00073-7
Cai Z, Guldbrandtsen B, Lund MS, Sahana G (2018) Prioritizing candidate genes post-GWAS using multiple sources of data for mastitis resistance in dairy cattle. BMC Genom 19:656. https://doi.org/10.1186/s12864-018-5050-x
Cai Z, Dusza M, Guldbrandtsen B, Lund MS, Sahana G (2020) Distinguishing pleiotropy from linked QTL between milk production traits and mastitis resistance in Nordic Holstein cattle. Genet Sel Evol 52:19. https://doi.org/10.1186/s12711-020-00538-6
Carlén E, Strandberg E, Roth A (2004) Genetic parameters for clinical mastitis, somatic cell score, and production in the first three lactations of Swedish Holstein cows. J Dairy Sci 87(9):3062–3070. https://doi.org/10.3168/jds.S0022-0302(04)73439-6
Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. https://doi.org/10.1186/s13742-015-0047-8
Chen X, Cheng Z, Zhang S, Werling D, Cheng Z (2015) Combining genome wide association studies and differential gene expression data analyses identifies candidate genes affecting mastitis caused by two different pathogens in the dairy cow. Open J Anim Sci 5:358–393. https://doi.org/10.4236/ojas.2015.54040
Chen X, Chen S, Li Y, Gao Y, Huang S, Li H, Zhu Y (2019) SMURF1-mediated ubiquitination of ARHGAP26 promotes ovarian cancer cell invasion and migration. Exp Mol Med 51(4):1–2. https://doi.org/10.1038/s12276-019-0236-0
Crowl JT, Stetson DB (2018) SUMO2 and SUMO3 redundantly prevent a noncanonical type I interferon response. Proc Natl Acad Sci 115(26):6798–6803. https://doi.org/10.1073/pnas.1802114115
Das S, Sarkar A, Choudhury SS, Owen KA, Castillo V, Fox S, Eckmann L, Elliott MR, Casanova JE, Ernst PB (2015) ELMO1 has an essential role in the internalization of Salmonella Typhimurium into enteric macrophages that impacts disease outcome. Cell Mol Gastroenterol Hepatol 1(3):311–324. https://doi.org/10.1016/j.jcmgh.2015.02.003
Di X, Jin X, Ma H, Wang R, Cong S, Tian C, Liu J, Zhao M, Li R, Wang K (2019) The oncogene IARS2 promotes non-small cell lung cancer tumorigenesis by activating the AKT/MTOR pathway. Front Oncol 9:393. https://doi.org/10.3389/fonc.2019.00393
Eichler TW, Totland C, Haugen M, Qvale TH, Mazengia K, Storstein A, Haukanes BI, Vedeler CA (2013) CDR2L antibodies: a new player in paraneoplastic cerebellar degeneration. PLoS One 8(6):e66002. https://doi.org/10.1371/journal.pone.0066002
Fadista J, Manning AK, Florez JC, Groop L (2016) The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur J Hum Genet 24(8):1202–1205. https://doi.org/10.1038/ejhg.2015.269
Fortune M, Guo H, Burren O, Schofield E, Walker NM, Ban M, Sawcer SJ, Bowes J, Worthington J, Barton A, Eyre S, Todd JA, Wallace C (2015) Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat Genet 47:839–846. https://doi.org/10.1038/ng.3330
Gao H, Lund MS, Zhang Y, Su G (2013) Accuracy of genomic prediction using different models and response variables in the Nordic Red cattle population. J Anim Breed Genet 130(5):333–340. https://doi.org/10.1111/jbg.12039
Guo G, Lund MS, Zhang Y, Su G (2010) Comparison between genomic predictions using daughter yield deviation and conventional estimated breeding value as response variables. J Anim Breed Genet 127(6):423–432. https://doi.org/10.1111/j.1439-0388.2010.00878.x
Hadfield JD (2010) MCMC Methods for Multi-Response Generalized Linear Mixed Models: The MCMCglmm R Package. J Stat Softw 33(2):1–22. http://www.jstatsoft.org/v33/i02/
Halasa T, Huijps K, Østerås O, Hogeveen H (2007) Economic effects of bovine mastitis and mastitis management: a review. Vet Q 29:18–31. https://doi.org/10.1080/01652176.2007.9695224
Heringstad B, Klemetsdal G, Ruane J (2000) Selection for mastitis resistance in dairy cattle: a review with focus on the situation in the Nordic countries. Livest Prod Sci 64:95–106. https://doi.org/10.1016/S0301-6226(99)00128-1
Heringstad B, Chang YM, Gianola D, Klemetsdal G (2004) Multivariate threshold model analysis of clinical mastitis in multiparous Norwegian Dairy Cattle. J Dairy Sci 87(9):3038–3046. https://doi.org/10.3168/jds.S0022-0302(04)73436-0
http://cpcsea.nic.in/Content/55_1_GUIDELINES.aspx. Accessed on 08 Apr 2020
https://genome.ucsc.edu/. Accessed 10 Apr 2020
https://github.com/sahirbhatnagar/manhattanly/. Accessed 10 Apr 2020
https://www.animalgenome.org/cgi-bin/QTLdb/BT/index. Accessed 01 Sept 2020
https://www.dairyknowledge.in/sites/default/files/genomic_selection.pdf. Accessed 09 Apr 2020
https://www.nddb.coop/services/animalbreeding/geneticimprovement/genomic. Accessed 09 Apr 2020
Jha A, Singh AK, Weissgerber P, Freichel M, Flockerzi V, Flavell RA, Jha MK (2015) Essential roles for Cavβ2 and Cav1 channels in thymocyte development and T cell homeostasis. Sci Signal 8(399):103. https://doi.org/10.1126/scisignal.aac7538
Jiang L, Liu J, Sun D, Ma P, Ding X, Yu Y, Zhang Q (2010) Genome wide association studies for milk production traits in Chinese Holstein population. PLoS One 5(10):e13661. https://doi.org/10.1371/journal.pone.0013661
Jiang G, Tan Y, Wang H, Peng L, Chen HT, Meng XJ, Li LL, Liu Y, Li WF, Shan H (2019) The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol Cancer 18:17. https://doi.org/10.1186/s12943-019-0944-z
Johansson K, Eriksson S, Pösö J, Toivonen M, Nielsen US, Eriksson JA, Aamand GP (2006) Genetic evaluation of udder health traits for Denmark, Finland and Sweden. Interbull Bull 35:92–96
Ju Z, Jiang Q, Wang J, Wang X, Yang C, Sun Y, Zhang Y, Wang C, Gao Y, Wei X, Hou M, Huang J (2020) Genome-wide methylation and transcriptome of blood neutrophils reveal the roles of DNA methylation in affecting transcription of protein-coding genes and miRNAs in E. coli-infected mastitis cows. BMC Genom 21:102. https://doi.org/10.1186/s12864-020-6526-z
Karatzias H, Karatzia M, Panagiotis K (2013) Occurrence, etiology and prevention of abomasal displacement in dairy. Cattle: domestication, diseases and the environment. Nova Science Publishers Inc., New York, pp 127–138
Kirsanova E, Heringstad B, Lewandowska-Sabat A, Olsaker I (2020) Identification of candidate genes affecting chronic subclinical mastitis in Norwegian Red cattle: combining genome-wide association study, topologically associated domains and pathway enrichment analysis. Anim Genet 51(1):22–31. https://doi.org/10.1111/age.12886
Koeck A, Heringstad B, Egger-Danner C, Fuerst C, Winter P (2010) Genetic analysis of clinical mastitis and somatic cell count traits in Austrian Fleckvieh cows. J Dairy Sci 93(12):5987–5995. https://doi.org/10.3168/jds.2010-3451
Krömker V, Leimbach S (2017) Mastitis treatment—reduction in antibiotic usage in dairy cows. Reprod Dom Anim 52(Suppl. 3):21–29. https://doi.org/10.1111/rda.13032
Kurz P, Yang Z, Weiss RB, Wilson DJ, Rood KA, Liu GE, Wang Z (2019) A genome-wide association study for mastitis resistance in phenotypically well-characterized Holstein dairy cattle using a selective genotyping approach. Immunogenetics 71(1):35–47. https://doi.org/10.1007/s00251-018-1088-9
Lee HJ, Kim JM, Kim KH, Heo JI, Kwak SJ, Han JA (2015) Genotoxic stress/p53-induced DNAJB9 inhibits the pro-apoptotic function of p53. Cell Death Differ 22(1):86–95. https://doi.org/10.1038/cdd.2014.116
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28(18):2397–2399. https://doi.org/10.1093/bioinformatics/bts444
Lund MS, Guldbrandtsen B, Buitenhuis AJ, Thomsen B, Bendixen C (2008) Detection of quantitative trait loci in Danish Holstein cattle affecting clinical mastitis, somatic cell score, udder conformation traits, and assessment of associated effects on milk yield. J Dairy Sci 91(10):4028–4036. https://doi.org/10.3168/jds.2007-0290
Marete A, Sahana G, Fritz S, Lefebvre R, Barbat A, Lund MS, Guldbrandtsen B, Boichard D (2018) Genome-wide association study for milking speed in French Holstein cows. J Dairy Sci 101(7):6205–6219. https://doi.org/10.3168/jds.2017-14067
Márquez A, Kerick M, Zhernakova A, Gutierrez-Achury J, Chen WM, Onengut-Gumuscu S, González-Álvaro I, Rodriguez-Rodriguez L, Rios-Fernández R, González-Gay MA, Coeliac Disease Immunochip Consortium, Rheumatoid Arthritis Consortium International for Immunochip (RACI), International Scleroderma Group, Type 1 Diabetes Genetics Consortium, Mayes MD, RayChaudhari S, Rich SS, Wijmenga C, Martin J (2018) Meta-analysis of Immunochip data of four autoimmune diseases reveals novel single disease and cross-phenotype associations. Genome Med 10:97. https://doi.org/10.1186/s13073-018-0604-8
Meredith BK, Kearney FJ, Finlay EK, Bradley DG, Fahey AG, Berry DP, Lynn DJ (2012) Genome-wide associations for milk production and somatic cell score in Holstein–Friesian cattle in Ireland. BMC Genet 13:21. https://doi.org/10.1186/1471-2156-13-21
Meredith BK, Berry DP, Kearney F, Finlay EK, Fahey AG, Bradley DG, Lynn DJ (2013) A genome-wide association study for somatic cell score using the Illumina high-density bovine beadchip identifies several novel QTL potentially related to mastitis susceptibility. Front Genet 4:229. https://doi.org/10.3389/fgene.2013.00229
Mömke S, Sickinger M, Lichtner P, Doll K, Rehage J, Distl O (2013) Genome-wide association analysis identifies loci for left-sided displacement of the abomasum in German Holstein cattle. J Dairy Sci 96(6):3959–3964. https://doi.org/10.3168/jds.2012-5679
Muncie SA, Cassady JP, Ashwell MS (2006) Refinement of quantitative trait loci on bovine chromosome 18 affecting health and reproduction in US Holsteins. Anim Genet 37(3):273–275. https://doi.org/10.1111/j.1365-2052.2006.01425.x
Nagy G, Ward J, Mosser DD, Koncz A, Gergely P Jr, Stancato C, Qian Y, Fernandez D, Niland B, Grossman CE et al (2006) Regulation of CD4 expression via recycling by HRES-1/RAB4 controls susceptibility to HIV infection. J Biol Chem 281:34574–34591. https://doi.org/10.1074/jbc.m606301200
Nash DL, Rogers GW, Cooper JB, Hargrove GL, Keown JF, Hansen LB (2000) Heritability of clinical mastitis incidence and relationships with sire transmitting abilities for somatic cell score, udder type traits, productive life, and protein yield. J Dairy Sci 83(10):2350–2360. https://doi.org/10.3168/jds.S0022-0302(00)75123-X
Nofrini V, Giacomo DD, Mecucci C (2016) Nucleoporin genes in human diseases. Eur J Hum Genet 24(10):1388–1395. https://doi.org/10.1038/ejhg.2016.25
Ogorevc J, Kunej T, Razpet A, Dovc P (2009) Database of cattle candidate genes and genetic markers for milk production and mastitis. Anim Genet 40(6):832–851. https://doi.org/10.1111/j.1365-2052.2009.01921.x
Oliveira HR, Lourenco D, Masuda Y, Misztal I, Tsuruta S, Jamrozik J, Brito LF, Silva FF, Cant JP, Schenkel FS (2019) Single-step genome-wide association for longitudinal traits of Canadian Ayrshire, Holstein, and Jersey dairy cattle. J Dairy Sci 102(11):9995–10011. https://doi.org/10.3168/jds.2019-16821
Olsen HG, Knutsen TM, Lewandowska-Sabat AM, Grove H, Nome T, Svendsen M, Arnyasi M, Sodeland M, Sundsaasen KK, Dahl SR et al (2016) Fine mapping of a QTL on bovine chromosome 6 using imputed full sequence data suggests a key role for the group-specific component (GC) gene in clinical mastitis and milk production. Genet Sel Evol 48(1):79. https://doi.org/10.1186/s12711-016-0257-2
Pisanu S, Cacciotto C, Pagnozzi D, Uzzau S, Pollera C, Penati M, Bronzo V, Addis MF (2020) Impact of Staphylococcus aureus infection on the late lactation goat milk proteome: new perspectives for monitoring and understanding mastitis in dairy goats. J Proteom 221:103763. https://doi.org/10.1016/j.jprot.2020.103763
Prasad AS (2008) Zinc in human health: effect of Zinc on immune cells. Mol Med 14(5–6):353–357. https://doi.org/10.2119/2008-00033.Prasad
Preisler MT, Weber PS, Tempelman RJ, Erskine RJ, Hunt H, Burton JL (2000) Glucocorticoid receptor down-regulation in neutrophils of periparturient cows. Am J Vet Res 61:14–19. https://doi.org/10.2460/ajvr.2000.61.14
Qanbari S (2020) On the extent of linkage disequilibrium in the genome of farm animals. Front Genet 10:1304. https://doi.org/10.3389/fgene.2019.01304
Radtke D, Lacher SM, Szumilas N, Sandrock L, Ackermann J, Nitschke L, Zinser E (2016) Grb2 is important for T cell development, Th cell differentiation, and induction of experimental autoimmune encephalomyelitis. J Immunol 196(7):2995–3005. https://doi.org/10.4049/jimmunol.1501764M
Rinaldi M, Li RW, Bannerman DD, Daniels KM, Evock-Clover C, Silva MV, Paape MJ, Van Ryssen B, Burvenich C, Capuco AV (2010) A sentinel function for teat tissues in dairy cows: dominant innate immune response elements define early response to E. coli mastitis. Funct Integr Genom 10(1):21–38. https://doi.org/10.1007/s10142-009-0133-z
Rupp R, Boichard D (2003) Genetics of resistance to mastitis in dairy cattle. Vet Res 34(5):671–688. https://doi.org/10.1051/vetres:2003020
Sahana G, Guldbrandtsen B, Thomsen B, Holm LE, Panitz F, Brøndum RF, Bendixen C, Lund MS (2014) Genome-wide association study using high-density single nucleotide polymorphism arrays and whole-genome sequences for clinical mastitis traits in dairy cattle. J Dairy Sci 97(11):7258–7275. https://doi.org/10.3168/jds.2014-8141
Sambrook J, Russel DW (2006) Rapid isolation of yeast DNA. CSH Protoc. https://doi.org/10.1101/pdb.prot4039
Schmid M, Bennewitz J (2017) Invited review: genome-wide association analysis for quantitative traits in livestock—a selective review of statistical models and experimental designs. Arch Anim Breed 60:335–346. https://doi.org/10.5194/aab-60-335-2017
Schrooten C, Bovenhuis H, Coppieters W, Van Arendonk JA (2000) Whole genome scan to detect quantitative trait loci for conformation and functional traits in dairy cattle. J Dairy Sci 83(4):795–806. https://doi.org/10.3168/jds.S0022-0302(00)74942-3
Schubert M, Panja D, Haugen M, Bramham CR, Vedeler CA (2014) Paraneoplastic CDR2 and CDR2L antibodies affect Purkinje cell calcium homeostasis. Acta Neuropathol 128:835–852. https://doi.org/10.1007/s00401-014-1351-6
Sentitula YBR, Kumar R (2012) Incidence of staphylococci and streptococci during winter in mastitic milk of Sahiwal cow and Murrah buffaloes. Indian J Microbiol 52(2):153–159. https://doi.org/10.1007/s12088-011-0207-1
Sharma C, Rokana N, Chandra M, Singh BP, Gulhane RD, Gill J, Ray P, Puniya AK, Panwar H (2018) Antimicrobial resistance: its surveillance, impact, and alternative management strategies in dairy animals. Front Vet Sci 4:237. https://doi.org/10.3389/fvets.2017.00237
Shook GE (1993) Genetic improvement of mastitis through selection on somatic cell count. Vet Clin N Am Food Anim Pract 9(3):563–581. https://doi.org/10.1016/s0749-0720(15)30622-8
Siebert LJ (2017) Identifying genome associations with unique mastitis phenotypes in response to intramammary Streptococcus uberis challenge. Ph.D. Dissertation, University of Tennessee, Tennessee, US. https://trace.tennessee.edu/utk_graddiss/4424
Singh RS, Bansal BK, Gupta DK (2014) Udder health in relation to udder and teat morphometry in Holstein Friesian × Sahiwal crossbred dairy cows. Trop Anim Health Prod 46:93–98. https://doi.org/10.1007/s11250-013-0454-8
Sinha R, Sinha B, Kumari R, Vineeth MR, Verma A, Gupta ID (2019) Effect of season, stage of lactation, parity and levels of milk production on incidence of clinical mastitis in Karan Fries and Sahiwal cattle. Biol Rhythm Res. https://doi.org/10.1080/09291016.2019.1621064
Tal-Stein R, Fontanesi L, Dolezal M, Scotti E, Bagnato A, Russo V, Canavesi F, Friedmann A, Soller M, Lipkin E (2010) A genome scan for quantitative trait loci affecting milk somatic cell score in Israeli and Italian Holstein cows by means of selective DNA pooling with single- and multiple-marker mapping. J Dairy Sci 93(10):4913–4927. https://doi.org/10.3168/jds.2010-3254
Thompson-Crispi KA, Sargolzaei M, Ventura R, Abo-Ismail M, Miglior F, Schenkel F, Mallard BA (2014) A genome-wide association study of immune response traits in Canadian Holstein cattle. BMC Genom 15(1):559. https://doi.org/10.1186/1471-2164-15-559
Tiezzi F, Parker-Gaddis KL, Cole JB, Clay JS, Maltecca C (2015) A genome-wide association study for clinical mastitis in first parity US Holstein cows using single-step approach and genomic matrix re-weighting procedure. PLoS One 10(2):e0114919. https://doi.org/10.1371/journal.pone.0114919
Toda E, Terashima Y, Esaki K, Yoshinaga S, Sugihara M, Kofuku Y, Shimada I, Suwa M, Kanegasaki S, Terasawa H, Matsushima K (2014) Identification of a binding element for the cytoplasmic regulator FROUNT in the membrane-proximal C-terminal region of chemokine receptors CCR2 and CCR5. Biochem J 457(2):313–322. https://doi.org/10.1042/BJ20130827
Urioste JI, Franzén J, Windig JJ, Strandberg E (2012) Genetic relationships among mastitis and alternative somatic cell count traits in the first 3 lactations of Swedish Holsteins. J Dairy Sci 95(6):3428–3434. https://doi.org/10.3168/jds.2011-4739
Van de Schoot R, Broere JJ, Perryck KH, Zwijnenburgm MZ, Loey NEV (2015) Analyzing small data sets using Bayesian estimation: the case of posttraumatic stress symptoms following mechanical ventilation in burn survivors. Eur J Psychotraumatol. https://doi.org/10.3402/ejpt.v6.25216.10.3402/ejpt.v6.25216
Vanshylla K, Bartsch C, Hitzing C, Krümpelmann L, Wienands J, Engels N (2018) Grb2 and GRAP connect the B cell antigen receptor to Erk MAP kinase activation in human B cells. Sci Rep 8:4244. https://doi.org/10.1038/s41598-018-22544-x
Villemereuil P (2019) On the relevance of Bayesian statistics and MCMC for animal models. J Anim Breed Genet 136(5):339–340. https://doi.org/10.1111/jbg.12426
Wang X, Ma P, Liu J, Zhang Q, Zhang Y, Ding X, Jiang L, Wang Y, Zhang Y, Sun D, Zhang S, Su G, Yu Y (2015) Genome-wide association study in Chinese Holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet 16:111. https://doi.org/10.1186/s12863-015-0263-3
Weigel KA, Shook GE (2018) Genetic selection for mastitis resistance. Vet Clin N Am Food Anim Pract 34(3):457–472. https://doi.org/10.1016/j.cvfa.2018.07.001
Welderufael BG, Løvendahl P, de Koning DJ, Janss LLG, Fikse WF (2018) Genome-wide association study for susceptibility to and recoverability from mastitis in Danish Holstein cows. Front Genet 9:141. https://doi.org/10.3389/fgene.2018.00141
Wellenberg GJ, van der Poel WH, Van Oirschot JT (2002) Viral infections and bovine mastitis: a review. Vet Microbiol 88(1):27–45. https://doi.org/10.1016/s0378-1135(02)00098-6
Wu X, Lund MS, Sahana G, Guldbrandtsen B, Sun D, Zhang Q, Su G (2015) Association analysis for udder health based on SNP-panel and sequence data in Danish Holsteins. Genet Sel Evol 47:50. https://doi.org/10.1186/s12711-015-0129-1
www.cog-genomics.org/plink/1.9/ Purcell S, Chang C PLINK 1.9. Accessed 09 Apr 2020
Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, Outtz H, Kitajewski J, Shi C, Weber S et al (2012) Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol 13(7):642–650. https://doi.org/10.1038/ni.2304
Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, Smith AV, Ingelsson E, O’Connell JR, Mangino M et al (2011) Genomic inflation factors under polygenic inheritance. Eur J Hum Genet 19:807–812. https://doi.org/10.1038/ejhg.2011.39
Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, Cui J, Zeng YX, Li J (2014) Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget 5(14):5439–5452
Zastre JA, Hanberry BS, Sweet RL, McGinnis AC, Venuti KR, Bartlett MG, Govindarajan R (2013) Up-regulation of vitamin B1 homeostasis genes in breast cancer. J Nutr Biochem 24(9):1616–1624. https://doi.org/10.1016/j.jnutbio.2013.02.002
Zhang Z, Ersoz E, Lai C, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM et al (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355–360. https://doi.org/10.1038/ng.546
Zhang L, Boeren S, Van Hooijdonk ACM, Vervoort JM, Hettinga KA (2015) A proteomic perspective on the changes in milk proteins due to high somatic cell count. J Dairy Sci 98:5339–5351. https://doi.org/10.3168/jds.2014-9279
Zhang H, Jiang H, Fan Y, Chen Z, Li M, Mao Y, Karrow NA, Loor JJ, Moore S, Yang Z (2018) Transcriptomics and iTRAQ-proteomics analyses of bovine mammary tissue with Streptococcus agalactiae-induced mastitis. J Agric Food Chem 66(42):11188–11196. https://doi.org/10.1021/acs.jafc.8b02386
Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS et al (2009) A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol 10(4):R42–R72. https://doi.org/10.1186/gb-2009-10-4-r42
Zwald NR, Weigel KA, Chang YM, Welper RD, Clay JS (2006) Genetic analysis of clinical mastitis data from on-farm management software using threshold models. J Dairy Sci 89:330–336. https://doi.org/10.3168/jds.S0022-0302(06)72098-7
Acknowledgements
The authors are grateful to Director, National Dairy Research Institute (NDRI) for providing necessary support and staff of Livestock Health complex and Livestock Research Centre for facilitating sample collection and data recording. We are indebted to National Dairy Development Board (NDDB), Anand, Gujarat, for genotyping the samples free of cost, as a part of MoU between NDRI and NDDB. The first author is also thankful to Dr. S.K. Onteru for helping in the interpretation of results and Indian Council of Medical Research (ICMR) for providing Junior Research Fellowship (JRF).
Funding
No financial grant was received to carry out this study.
Author information
Authors and Affiliations
Contributions
AK and SMD conceived and designed the study, AK and NN performed the statistical analysis, NN and SKN helped in laboratory work and genotyping of samples, VSR and VY facilitated data curation, and AK and SMD wrote the paper.
Corresponding author
Ethics declarations
Conflict of interests
All the authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Kour, A., Deb, S.M., Nayee, N. et al. Understanding the genomic architecture of clinical mastitis in Bos indicus. 3 Biotech 11, 466 (2021). https://doi.org/10.1007/s13205-021-03012-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-03012-2