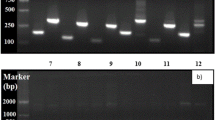
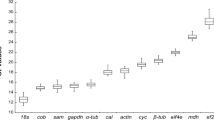
Abstract
The accurate measurement of gene expression via quantitative real-time reverse transcription PCR (qRT-PCR) heavily relies on the choice of valid reference gene(s) for data normalization. Resting cyst is the dormant stage in the life cycle of dinoflagellate, which plays crucial roles in HAB-forming dinoflagellate ecology. However, only limited investigations have been conducted on the reference gene selection in dinoflagellates. Gap remained in our knowledge about appropriate HKGs for normalizing gene expression in different life stages, which laid obstacles for the application of qRT-PCR to the HAB-forming group. In this study, six candidate reference genes, 18S ribosomal RNA (18S), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), α-tubulin (TUA), β-tubulin (TUB), actin (ACT) and cytochrome oxidase subunit 1 (COX1), were evaluated for their expression stability with qRT-PCR and three statistical algorithms (GeNorm, NormFinder, and BestKeeper) for the cosmopolitan, harmful algal bloom-forming dinoflagellate Akashiwo sanguinea. Expression patterns were observed across 18 biological samples, including cells at resting stages (resting cysts), different growth stages, in darkness, exposed to abscisic acid (ABA) and exposed to temperature stress. The results indicated that TUA, 18S and GAPDH were relatively stable across all tested scenarios. While the best-recommended reference genes differed across experimental groups, the pairs of ACT and TUA, 18S and GAPDH were the most reliable for cells at different growth stages and darkness treatment. The combination of TUA and TUB was the best choice for normalization in resting cysts and in ABA treatment, respectively. The pair of ACT and COX1 was suitable for temperature treatments. This study was the first to investigate the stable internal reference genes in dinoflagellates at different stages of life cycle, particularly in resting cysts. Our results provided useful information for selection of reference genes in dinoflagellates regarding quantification of gene expression at different experimental scenarios, which will facilitate more accurate and widespread use of qRT-PCR in gene analysis of dinoflagellates and help to design primers targeting orthologous genes in other algal species.
Similar content being viewed by others
References
Andersen C L, Jensen J L, Ørntoft T F. 2004. Normalization of realtime quantitative reverse transcription-PCR data: a modelbased variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res, 64(15): 5245–5250
Anderson D M, Cembella A D, Hallegraeff G M. 2012. Progress in understanding harmful algal blooms: Paradigm shifts and new technologies for research, monitoring, and management. Annu Rev Mar Sci, 4(1): 143–176
Bas A, Forsberg G, Hammarström S, et al. 2004. Utility of the housekeeping genes 18S rRNA, ß-actin and glyceraldehyde-3-phosphate- dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol, 59(6): 566–573
Boldt L, Yellowlees D, Leggat W. 2010. Measuring Symbiodinium sp. gene expression patterns with quantitative real-time PCR. In: Proceedings of the 11th International Coral Reef Symposium. Florida: Ft Lauderdale, 118–122
Botes L, Smit A J, Cook P A. 2003. The potential threat of algal blooms to the abalone (Haliotis midae) mariculture industry situated around the South African coast. Harmful Algae, 2(4): 247–259
Bravo I, Figueroa R I. 2014. Towards an ecological understanding of dinoflagellate cyst functions. Microorganisms, 2(1): 11–32
Brunner A M, Yakovlev I A, Strauss S H. 2004. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol, 4: 14
de Almeida M R, Ruedell C M, Ricachenevsky F K, et al. 2010. Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill. BMC Mol Biol, 11: 73
Demidenko N V, Logacheva M D, Penin A A. 2011. Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS One, 6(5): e19434
Deng Yunyan, Yao Jianting, Wang Xiuliang, et al. 2012. Transcriptome sequencing and comparative analysis of Saccharina japonica (Laminariales, Phaeophyceae) under blue light induction. PLoS One, 7(6): e39704
Doblin M A, Blackburn S I, Hallegraeff G M. 1999. Growth and biomass stimulation of the toxic dinoflagellate Gymnodinium catenatum (Graham) by dissolved organic substances. J Exp Mar Biol Ecol, 236(1): 33–47
Elbrächter M. 2003. Dinophyte reproduction: progress and conflicts. J Phycol, 39(4): 629–632
Expósito-Rodríguez M, Borges A A, Borges-Pérez A, et al. 2008. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol, 8: 131
Guillard R R L. 1975. Culture of phytoplankton for feeding marine invertebrates. In: Smith W L, Chanley M H, eds. Culture of Marine Invertebrate Animals. New York: Plenum Press
Guo Ruoyu, Ki J S. 2012. Evaluation and validation of internal control genes for studying gene expression in the dinoflagellate Prorocentrum minimum using real-time PCR. Eur J Protistol, 48(3): 199–206
Hallegraeff G M, Bolch C J. 1991. Transport of toxic dinoflagellate cysts via ships’ ballast water. Mar Pollut Bull, 22(1): 27–30
Hu Ruibo, Fan Chengming, Li Hongyu, et al. 2009. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol, 10: 93
Huggett J, Dheda K, Bustin S, et al. 2005. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun, 6(4): 279–284
Jessup D A, Miller M A, Ryan J P, et al. 2009. Mass stranding of marine birds caused by a surfactant-producing red tide. PLoS One, 4(2): e4550
Lee J M, Roche J R, Donaghy D J, et al. 2010. Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol Biol, 11: 8
Matsubara T, Nagasoe S, Yamasaki Y, et al. 2007. Effects of temperature, salinity, and irradiance on the growth of the dinoflagellate Akashiwo sanguinea. J Exp Mar Biol Ecol, 342(2): 226–230
Pfaffl M W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res, 29(9): e45
Pfaffl M W, Tichopad A, Prgomet C, et al. 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pairwise correlations. Biotechnol Lett, 26(6): 509–515
Radonic A, Thulke S, Mackay I M, et al. 2004. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun, 313(4): 856–862
Rosic N N, Pernice M, Rodriguez-Lanetty M, et al. 2011. Validation of housekeeping genes for gene expression studies in Symbiodinium exposed to thermal and light stress. Mar Biotechnol, 13(3): 355–365
Steidinger K A, Tangen K. 1996. Dinoflagellates. In: Tomas C R, ed. Identifying Marine Diatoms and Dinoflagellates. New York: Academic Press
Tang Yingzhong, Gobler C J. 2012. The toxic dinoflagellate Cochlodinium polykrikoides (Dinophyceae) produces resting cysts. Harmful Algae, 20: 71–80
Tang Yingzhong, Gobler C J. 2015. Sexual resting cyst production by the dinoflagellate Akashiwo sanguinea: a potential mechanism contributing to the ubiquitous distribution of a harmful alga. J Phycol, 51(2): 298–309
Vandesompele J, De Preter K, Pattyn F, et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol, 3(7): research0034.1
Woelke C E. 1961. Pacific oyster Crassostrea gigas mortalities: with notes on common oyster predators in Washington waters. Proc Natl Shellfisheries Assoc, 50: 53–66
Author information
Authors and Affiliations
Corresponding author
Additional information
Foundation item: The National Natural Science Foundation of China-Shandong Joint Fund for Marine Science Research Centers under contract No. U1406403; the State Key Program of National Natural Science of China under contract No. 61533011; China Postdoctoral Science Foundation under contract Nos 2014M551969 and 2015T80754.
Rights and permissions
About this article
Cite this article
Deng, Y., Hu, Z., Ma, Z. et al. Validation of reference genes for gene expression studies in the dinoflagellate Akashiwo sanguinea by quantitative real-time RT-PCR. Acta Oceanol. Sin. 35, 106–113 (2016). https://doi.org/10.1007/s13131-016-0887-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13131-016-0887-9