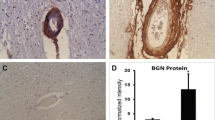
Abstract
Vascular smooth muscle cells (SMCs) undergo a series of dramatic changes in CADASIL, the most common inherited cause of vascular dementia and stroke. NOTCH3 protein accumulates and aggregates early in CADASIL, followed by loss of mature SMCs from the media of brain arteries and marked intimal proliferation. Similar intimal thickening is seen in peripheral arterial disease, which features pathological intimal cells including proliferative, dedifferentiated, smooth muscle-like cells deficient in SMC markers. Limited studies have been performed to investigate the differentiation state and location of SMCs in brain vascular disorders. Thus, we investigated the distribution of cells expressing SMC markers in a group of genetically characterized, North American CADASIL brains. We quantified brain RNA abundance of these markers in nine genetically verified cases of CADASIL and found that mRNA expression for several mature SMC markers was increased in CADASIL brain compared to age-matched control. Immunohistochemical studies and in situ hybridization localization of mRNA demonstrated loss of SMCs from the arterial media, and SMC marker-expressing cells were instead redistributed into the intima of diseased arteries and around balloon cells of the degenerating media. We conclude that, despite loss of medial smooth muscle cells in diseased arteries, smooth muscle markers are not lost from CADASIL brain, but rather, the localization of cells expressing mature SMC markers changes dramatically.






Similar content being viewed by others
References
Miao Q, Paloneva T, Tuominen S, Pöyhönen M, Tuisku S, Viitanen M, et al. Fibrosis and stenosis of the long penetrating cerebral arteries: the cause of the white matter pathology in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Brain Pathol. 2004;14:358–64. https://doi.org/10.1111/j.1750-3639.2004.tb00078.x.
Kalimo H, Viitanen M, Amberla K, et al. CADASIL: hereditary disease of arteries causing brain infarcts and dementia. Neuropathol Appl Neurobiol. 1999;25:257–65. https://doi.org/10.1046/j.1365-2990.1999.00198.x.
Baudrimont M, Dubas F, Joutel A, Tournier-Lasserve E, Bousser M-G. Case report autosomal dominant leukoencephalopathy and subcortical ischemic stroke. Stroke. 1993;24:122–6.
Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssière C, et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet. 1997;350:1511–5. https://doi.org/10.1016/S0140-6736(97)08083-5.
Yamamoto Y, Craggs LJL, Watanabe A, Booth T, Attems J, Low RWC, et al. Brain microvascular accumulation and distribution of the NOTCH3 ectodomain and granular osmiophilic material in CADASIL. J Neuropathol Exp Neurol. 2013;72(5):416–31. https://doi.org/10.1097/NEN.0b013e31829020b5.
Ruchoux MM, Domenga V, Brulin P, Maciazek J, Limol S, Tournier-Lasserve E, et al. Transgenic mice expressing mutant Notch3 develop vascular alterations characteristic of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Am J Pathol. 2003;162(1):329–42. https://doi.org/10.1016/S0002-9440(10)63824-2.
Takahashi K, Adachi K, Yoshizaki K, Kunimoto S, Kalaria RN, Watanabe A. Mutations in NOTCH3 cause the formation and retention of aggregates in the endoplasmic reticulum, leading to impaired cell proliferation. Hum Mol Genet. 2009;19(1):79–89. https://doi.org/10.1093/hmg/ddp468.
Dong H, Ding H, Young K, Blaivas M, Christensen PJ, Wang MM. Advanced intimal hyperplasia without luminal narrowing of leptomeningeal arteries in CADASIL. Stroke. 2013;44(5):1456–8. https://doi.org/10.3174/ajnr.A1650.Side.
Varcoe RL, Mikhail M, Guiffre AK, Pennings G, Vicaretti M, Hawthorne WJ. The role of the fibrocyte in intimal hyperplasia. J Thromb Haemost. 2006;4:1125–33.
Ruchoux M-M, Maurage C-A. CADASIL: cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. J Neuropathol Exp Neurol. 1997;56(9):947–64.
Gutierrez-Molina M, Caminero Rodriguez A, Martinez Garcia C, Arpa Gutierrez J, Morales Bastos C, Amer G. Small arterial granular degeneration in familial Binswanger’s syndrome. Acta Neuropathol. 1994;87:98–105.
Ruchoux M-M, Guerouaou D, Vandenhaute B, Pruvo J-P, Vermersch P, Leys D. Systemic vascular smooth muscle cell impairment in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Acta Neuropathol. 1995;89:500–12.
Skalli O, Ropraz P, Trzeciak A, Benzonana G, Gillessen D, Gabbiani G. A monoclonal antibody against alpha-smooth muscle actin: a new probe for smooth muscle differentiation. J Cell Biol. 1986;103(6 Pt 2):2787–96. https://doi.org/10.1083/jcb.103.6.2787.
Brennan-Krohn T, Salloway S, Correia S, Dong M, De La Monte SM. Glial vascular degeneration in CADASIL. J Alzheimers Dis. 2010;21(4):1393–402. https://doi.org/10.3233/JAD-2010-100036.
Szpak GM, Lewandowska E, Wierzba-Bobrowicz T, et al. Small cerebral vessel disease in familial amyloid and non-amyloid angiopathies: FAD-PS-1 (P117L) mutation and CADASIL. Immunohistochemical and ultrastructural studies. Folia Neuropathol. 2007;45(4):192–204.
Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. https://doi.org/10.1152/physrev.00041.2003.
Frid MG, Printesva OY, Chiavegato A, Faggin E, Scatena M, Koteliansky VE, et al. Myosin heavy-chain isoform composition and distribution in developing and adult human aortic smooth muscle. J Vasc Res. 1993;30:279–92.
Madsen CS, Regan CP, Hungerford JE, White SL, Manabe I, Owens GK. Smooth muscle specific expression of the smooth muscle myosin heavy chain gene in transgenic mice requires 5′-flanking and first intronic DNA sequence. Circ Res. 1998;82:908–17. https://doi.org/10.1161/01.RES.82.8.908.
Miano JM, Cserjesi P, Ligon KL, Periasamy M, Olson EN. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circ Res. 1994;75:803–12. https://doi.org/10.1161/01.RES.75.5.803.
van der Loop FT, Schaart G, Timmer ED, Ramaekers FC, van Eys GJ. Smoothelin, a novel cytoskeletal protein specific for smooth muscle cells. J Cell Biol. 1996;134(2):401–11.
Duband JL, Gimona M, Scatena M, Sartore S, Small JV. Calponin and SM22 as differentiation markers of smooth muscle: spatiotemporal distribution during avian embryonic development. Differentiation. 1993;55:1–11. https://doi.org/10.1111/j.1432-0436.1993.tb00027.x.
Gabbiani G, Schmid E, Winter S, Chaponnier C, de Ckhastonay C, Vandekerckhove J, et al. Vascular smooth muscle cells differ from other smooth muscle cells: predominance of vimentin filaments and a specific alpha-type actin. Proc Natl Acad Sci U S A. 1981;78(1):298–302. https://doi.org/10.1073/pnas.78.1.298.
Owens GK, Thompson MM. Developmental changes in isoactin expression in rat aortic smooth muscle cells in vivo. Relationship between growth and cytodifferentiation. J Biol Chem. 1986;261(28):13373–80.
Li L, Miano JM, Cserjesi P, Olson EN. SM22α, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ Res. 1996;78(2):188 LP–195.
Han DKM, Liau G. Identification and characterization of developmentally regulated genes in vascular smooth muscle cells. Circ Res. 1992;71(3):711–9.
Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75(3):487–517.
Miano JM, Olson EN. Expression of the smooth muscle cell calponin gene marks the early cardiac and smooth muscle cell lineages during mouse embryogenesis [published erratum appears in J Biol Chem 1997 Oct 24;272(43):27492]. J Biol Chem. 1996;271(12):7095–103.
Zhang X, Lee SJ, Young MF, Wang MM. The small leucine-rich proteoglycan BGN accumulates in CADASIL and binds to NOTCH3. Transl Stroke Res. 2015;6(2):148–55. https://doi.org/10.1007/978-1-4939-2914-6.
Zhang X, Lee SJ, Young KZ, Josephson DA, Michael D, Wang MM. Latent NOTCH3 epitopes unmasked in CADASIL and regulated by protein redox state. Brain Res. 2014;1583:230–6. https://doi.org/10.1016/j.brainres.2014.08.018.Latent.
Shawber CJ, Kitajewski J. Notch function in the vasculature: insights from zebrafish, mouse and man. BioEssays. 2004;26(3):225–34. https://doi.org/10.1002/bies.20004.
Kalimo H, Ruchoux M-M, Viitanen M, Kalaria RN, et al. Brain Pathol. 2002;12:371–84. https://doi.org/10.1111/j.1750-3639.2002.tb00451.x.
Regan CP, Adam PJ, Madsen CS, Owens GK. Molecular mechanisms of decreased smooth muscle differentiation marker expression after vascular injury. J Clin Invest. 2000;106:1139–47.
Aikawa M, Rabkin E, Voglic SJ, Shing H, Nagai R, Schoen FJ, et al. Muscle cells expressing smooth muscle myosin heavy. Circ Res. 1998;83(10):1015–26.
Yoon NK, Awad A-W, Kalani MYS, Taussky P, Park MS. Stent technology in ischemic stroke. Neurosurg Focus. 2017;42(4):1–9. https://doi.org/10.3171/2017.1.FOCUS16507.
Rensen SSM, Doevendans PAFM, van Eys GJJM. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Netherlands Hear J. 2007;15:100–8. https://doi.org/10.1007/BF03085963.
Hao H, Gabbiani G, Bochaton-Piallat ML. Arterial smooth muscle cell heterogeneity: implications for atherosclerosis and restenosis development. Arterioscler Thromb Vasc Biol. 2003;23:1510–20. https://doi.org/10.1161/01.ATV.0000090130.85752.ED.
Beamish JA, He P, Kottke-Marchant K, Marchant RE. Molecular regulation of contractile smooth muscle cell phenotype: implications for vascular tissue engineering. Tissue Eng Part B Rev. 2010;16(5):467–91. https://doi.org/10.1089/ten.teb.2009.0630.
Zhang X, Meng H, Blaivas M, Rushing EJ, Moore BE, Schwartz J, et al. von Willebrand factor permeates small vessels in CADASIL and inhibits smooth muscle gene expression. Transl Stroke Res. 2012;3(1):138–45. https://doi.org/10.1007/s12975-011-0112-2.Von.
Tikka S, Ng YP, Di MG, et al. CADASIL mutations and shRNA silencing of NOTCH3 affect actin organization in cultured vascular smooth muscle cells. J Cereb Blood Flow Metab. 2013;32:2171–80. https://doi.org/10.1038/jcbfm.2012.123.
Borgers M, Schaper J, Schaper W. The origin of subendothelial cells in developing coronary collaterals. Virchows Arch Abt A Path Anat. 1973;358:281–94.
Imai H, Lee KJ, Lee SK, Lett KT, O’Neal RM, Thomas WA. Ultrastructural features of aortic cells in mitosis in control and cholesterol-fed swine. Lab Investig. 1970;23:401–15.
Yang P, Hong MS, Fu C, Schmit BM, Su Y, Berceli SA, et al. Pre-existing smooth muscle cells contribute to neointimal cell repopulation at an incidence varying widely among individual lesions. Surgery. 2016;159(2):602–12. https://doi.org/10.1021/acsnano.5b07425.Molecular.
Herring B, Hoggatt AM, Burlak C, Offermanns S. Previously differentiated medial vascular smooth muscle cells contribute to neointima formation following vascular injury. Vasc Cell. 2014;6:21. https://doi.org/10.1186/2045-824X-6-21.
Chamley-Campbell J, Campbell GR, Ross R. The smooth muscle cell in culture. Physiol Rev. 1979;59(1):1–61.
Sata M, Saiura A, Kunisato A, Tojo A, Okada S, Tokuhisa T, et al. Hematopoietic stem cells differentiate into vascular cells that participate in the pathogenesis of atherosclerosis. Nat Med. 2002;8(4):403–9. https://doi.org/10.1038/nm0402-403.
Tanaka K, Sata M, Hirata Y, Nagai R. Diverse contribution of bone marrow cells to neointimal hyperplasia after mechanical vascular injuries. Circ Res. 2003;93(8):783–90. https://doi.org/10.1161/01.RES.0000096651.13001.B4.
Tang Z, Wang A, Yuan F, Yan Z, Liu B, Chu JS, et al. Differentiation of multipotent vascular stem cells contributes to vascular diseases. Nat Commun. 2012;3(875):875. https://doi.org/10.1038/ncomms1867.
Hao H, Ropraz P, Verin V, et al. Heterogeneity of smooth muscle cell populations cultured from pig coronary artery. Atheroscler Thromb Vasc Biol. 2002;22:1093–9. https://doi.org/10.1161/01.ATV.0000022407.91111.E4.
Lee SJ, Zhang X, Wang MM. Vascular accumulation of the small leucine rich proteoglycan decorin in CADASIL. Neuroreport. 2014;25(13):1059–63. https://doi.org/10.1097/WNR.0000000000000230.Vascular.
Zhang X, Lee SJ, Young MF, Wang MM. The small leucine-rich proteoglycan BGN accumulates in CADASIL and binds to NOTCH3. Transl Stroke Res. 2015;6(2):148–55. https://doi.org/10.1007/978-1-4939-2914-6.
Dong H, Blaivas M, Wang MM. Bidirectional encroachment of collagen into the tunica media in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Brain Res. 2012;1456:64–71. https://doi.org/10.1038/jid.2014.371.
Acknowledgements
We appreciate the support of CADASIL Together We Have Hope and members of the international CADASIL community who made donations used in this study.
Funding
This study was funded by the National Institutes of Health (NS099783 and HL108842) and the U.S. Department of Veterans Affairs (BX000375 and BX003824).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Gatti, J.R., Zhang, X., Korcari, E. et al. Redistribution of Mature Smooth Muscle Markers in Brain Arteries in Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy. Transl. Stroke Res. 10, 160–169 (2019). https://doi.org/10.1007/s12975-018-0643-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12975-018-0643-x