Abstract
Purpose
This study aimed to investigate the impact of CC chemokine receptor 7 (CCR7) on the recruitment and polarization of tumor-associated macrophages (TAMs) in oral squamous cell carcinoma (OSCC).
Methods
We analyzed CCR7 expression pattern, clinicopathological significance, and its association with M2 macrophage infiltration in OSCC by bioinformatic methods. Small interfering RNA (siRNA) was utilized to silence CCR7 in OSCC cells. Conditioned media (CM) was harvested from transfected OSCC cells to establish a co-culture model of THP-1 derived macrophages and OSCC cells. Transwell assay and cell adhesion assay were performed to examine the effect of CCR7 on macrophages recruitment and adhesion. Cytoskeleton was labelled by phalloidin to observe macrophage morphological changes. Moreover, phenotypic alteration of macrophages was measured using quantitative real-time PCR (qRT-PCR), flow cytometry, and immunofluorescence (IF) staining. Ultimately, recombinant human CCL19 and CCL21 were added into the medium of THP-1 derived macrophages to explore their effects on polarization in vitro.
Results
In OSCC patients, the overexpression of CCR7 positively correlated with lymph node metastasis and M2 macrophage infiltration. Macrophage not only exhibited enhanced migration, invasion and adhesion abilities, but also appeared more spindle and branched in vitro when treated with CM from OSCC cells. However, these phenomena were abrogated with knockdown of CCR7. We also discovered that inhibition of CCR7 in OSCC cells suppressed TAMs polarization to an M2 phenotype. In addition, recombinant human CCL19 and CCL21 promoted macrophage M2-polarization in vitro.
Conclusion
CCR7 in OSCC cells promoted recruitment and M2-polarization of THP-1 derived macrophages in vitro by regulating production of CCL19 and CCL21.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Head and neck squamous cell carcinoma (HNSCC) is one of the most common cancer worldwide and is responsible for around half a million deaths globally every year [1]. Among the many types of HNSCC, oral squamous cell carcinoma (OSCC) is the most prevalent pathological form [2, 3]. OSCC is a major health threat and one of the leading causes of cancer death in many regions and countries [2,3,4]. The global incidence of OSCC is predicted to rise by 62% to 856,000 cases by 2035 [2]. Although substantial advances have been made in screening, diagnosis, and multidisciplinary treatment, the mortality of patients with OSCC has remained steadily high over the past few decades [5]. In this context, an in-depth understanding of the OSCC pathogenesis is urgently needed.
Tumor microenvironment (TME) is a delicate dynamic ecosystem containing cellular and non-cellular components that cancer cells depend for sustained growth, invasion, and metastasis [6]. The intricate interplay between tumor cells and surrounding TME cells leads to malignant evolution of oral premalignant lesions and adverse prognosis of OSCC patients [7, 8]. For example, Kujan et al. found significant positive associations between regulatory T cell (Treg)-related proteins (including FOXP3, TGF-β, IL-6, and IL-10) and the histopathological grade of oral epithelial disorders, implying a possible role of tumor-infiltrating immune cells play in evading the immune system and potentially driving malignant transformation [9]. Macrophages are phagocytic and antigen-presenting cells of innate immune system. Mirroring the Th1/Th2 paradigm, macrophages could be divided into two subgroups based on their degree of differentiation and functional capacity, which represent two extreme edges of a continuum of phenotypic range. The classically activated M1-type boosts an antitumor response, while the alternatively activated macrophage that displays anti-inflammatory characteristic is included in the M2 classification [10]. Tumor-associated macrophages (TAMs), which tend to present a M2 phenotype, are among the most abundant and decisive immune cells in the TME [11]. Compelling evidences have proposed that TAMs create an immunosuppressive milieu, exhibit increased pro-tumoral activities and affect virtually all aspects of tumor tissues [12,13,14]. In OSCC, TAMs have been found particularly higher enriched in the metastatic group than in the non-metastatic and control groups, implying a possibly role in facilitating tumor invasion and metastasis [15]. Similar results were also acquired from several systematic review, showing that elevated concentrations of CD163+ TAMs were associated with worse prognosis, and may serve as one of the most promising predictors of survival in OSCC patients [16,17,18]. Due to these findings, TAMs have been proposed as a potential therapeutic target in OSCC immunotherapy [19]. It should be noted that versatility and plasticity are hallmarks of macrophages [20]. A diverse array of signals generated from tumor cells could profoundly and differentially influence the polarization of TAMs [21]. Nevertheless, the recruitment processes and functional alterations of macrophages in OSCC are still poorly elucidated. There is, therefore, an imperative need to explore the underlying molecular mechanisms of macrophage differentiation in OSCC.
Chemokines are a group of small, secreted, chemotactic proteins with a multifaceted role in various physiological and pathological events, ranging from thymocyte development and leukocyte migration to lymphoid tissue formation [22, 23]. Based on the structural conservation of both cysteine residues and disulfide bonds, chemokines can be further classified into 4 major groups: CXC, CC, C and CX3C [22]. CC chemokine receptor 7 (CCR7) is one of the most prominent chemokines and belongs to the CC subgroup. It has been reported to abnormally express in diverse tumor types [24] and has been blamed for enhancing tumor migration, epithelial mesenchymal transition (EMT) and cancer stemness through combination with its two CC motif ligands: CC chemokine ligand 19 (CCL19) and CC chemokine ligand 21 (CCL21) [25, 26]. More importantly, we have demonstrated that CCR7 contributes to malignant biological behaviors of OSCC cells through various up- and downstream signaling molecules [27,28,29]. Whether CCR7 participates TAMs infiltration in OSCC, however, remains highly undetermined. Hence, we investigated the function of CCR7 on macrophage recruitment and polarization in OSCC.
2 Materials and methods
2.1 Bioinformatic analysis
Initially, the differential mRNA expression data of CCR7 in diverse human cancers were compared with their matched normal tissues via tumor immune estimation resource 2.0 (TIMER2.0) [30] to outline the expression pattern of CCR7. The full names of tumor abbreviations were supplied in Supplementary Table S1. We also selected 2 datasets (GSE25099 and GSE74530) containing OSCC samples and normal oral tissue from gene expression omnibus (GEO) database [31] to further explore CCR7 expression in OSCC. Differences in CCR7 expression and distribution at the protein level were evaluated utilizing immunohistochemistry staining data of two patients (IDs: 1505 and 298) from the Human Protein Atlas (HPA) database [32]. To analyze the association between CCR7 and clinicopathological features of HNSCC, RNA sequencing (RNA-Seq) data and clinical data of HNSCC patients were retrieved from the cancer genome atlas (TCGA).
Then, we investigate the correlation of CCR7 expression with overall immune infiltration degree in TCGA-HNSCC patients by generating Immunescore using ESTIMATE (Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data) algorithm [33]. Furthermore, we explored whether CCR7 expression can affect various immune cell infiltration levels in TCGA-HNSCC patients by extracting data from TIMER [34] and TIMER2.0 web server.
2.2 Reagents and antibodies
Dulbecco’s modified Eagle’s medium (DMEM), Roswell Park Memorial Institute medium (RPMI-1640), reduced serum media Opti-MEM®, β-mercaptoethanol, phosphate buffered saline (PBS), fetal bovine serum (FBS), penicillin and streptomycin were obtained from Gibco (Carlsbad, CA, USA). Phorbol-12-myristate-13 acetate (PMA), paraformaldehyde (PFA) and dimethylsulfoxide (DMSO) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Recombinant human CCL19 (cat No: 300-29B) and CCL21 (cat No: 300-35A) were purchased from PeproTech (Rocky Hill, NJ, USA). Antibodies used for flow cytometry, including human Fc-receptor blocking solution (cat No: 422301) and CD206-PE (cat No: 321105) were from BioLegend (San Diego, CA, USA). A mouse monoclonal anti‑CD163 antibody (cat No: sc‑33715) from Santa Cruz (Dallas, TX, USA) was utilized for immunofluorescence staining. Transfection agent INTERFERin® (cat No: 409-10) was obtained from Polyplus (SA, France). All other chemicals were classified as analytical grade reagents.
2.3 Cell lines and culture conditions
Human acute monocytic leukemia cell line THP-1 was purchased from the Cell Bank/Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China). Two well-characterized CCR7 high-expressed OSCC cell lines, PCI-4B and PCI-37B, were kindly donated by the University of Pittsburgh Cancer Institute (Pittsburgh, PA, USA) [35, 36]. THP-1 monocytes were cultured in RPMI-1640, while PCI-4B and PCI-37B cells were maintained in low glucose DMEM. All cells were supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. The medium of THP-1 was complemented with an additional 0.05 mM β-mercaptoethanol. All cells were incubated in a humidified atmosphere with 5% CO2 at 37 °C.
2.4 Cell transfection
Small interfering RNA (siRNA) sequence 5′-CTGGTCGTGTTGACCTATA-3′ targeting human CCR7 (siCCR7) and negative control siRNA (siNC) were purchased from Ribobio (Guangzhou, China). The transient transfection was conducted according to the manufacturer’s instructions. In brief, PCI-4B and PCI-37B cells were seeded the day before transfection to reach 30–50% confluency at the time of transfection. siRNA duplexes were diluted into reduced serum media Opti-MEM®. Then, add transfection reagent INTERFERin® into the siRNA solution, vortex-mixed, and incubated for 10 min at room temperature. Finally, INTERFERin®-siRNA complexes were added in complete fresh medium to incubate cells at 37 ℃. 24 h later, the transfected cells were collected for further experiments. The knockdown efficiency of siRNA was tested by quantitative real-time PCR assay.
2.5 Quantitative real-time PCR (qRT-PCR)
Quantitative real-time PCR (qRT-PCR) was carried out to evaluate the mRNA expression levels of CCR7 and macrophages-related genes in OSCC and THP-1 cells. Total RNA was isolated from cultured cells with TRIzol Reagent (TaKaRa, Kyoto, Japan). After examining the concentration and purity using a NanoDrop spectrophotometer (Thermo Scientific, MA, USA), RNA was reversely transcribed into cDNA using the PrimeScript RT reagent Kit (TaKaRa, Kyoto, Japan) according to the protocols recommended by the manufacturer. Subsequently, the cDNA was subjected to qRT-PCR detection using a TB Green® Premix Ex Taq™ II reagent Kit (TaKaRa, Kyoto, Japan) via an ABI QuantStudio3 Real-Time PCR System (Applied Biosystems, CA, USA). Housekeeping gene GADPH was used to normalize relative mRNA levels, calculated by the 2−ΔΔCT method. The primer sequences designed by Sangon Biotech (Shanghai, China) are presented in Table 1.
2.6 Preparation of conditioned medium
To obtain conditioned media (CM) of OSCC cell lines, PCI-4B and PCI-37B, when up to 80% confluence, were washed by PBS thrice and cultured for an additional 72 h with fresh RPMI-1640 medium without serum. Then, the cell-free supernatants were harvested, centrifuged at 1000 rpm for 5 min and collected as CM after being filtered using 0.22-μm polyvinylidene difluoride membrane filters. CM was aliquoted, stored immediately at − 80 ℃ and avoid repeated freeze/thaws until future experiments.
2.7 Transwell cell migration assay
Transwell cell migration assays were conducted to examine the migration ability of macrophages in response to the CM of OSCC cells. THP-1 was plated on the upper compartment of a 24-well Transwell chamber (8-μm pore diameter; Corning, NY, USA) at 1 × 106 cells/ml density in 200 μl fresh serum-deprived RPMI-1640 medium. Meanwhile, 800 μl CM or fresh RPMI-1640 medium alone as a blank control for a spontaneous migration were added to the lower chamber. After 24 h, the cell suspension in the upper chamber was aspirated. The migrated cells within the lower chamber were photographed by Nikon Eclipse Ts2R inverted fluorescence microscope (Nikon, Tokyo, Japan) and then counted using LUNA-II™ Automated Cell Counter (Logos Biosystems, South Korea) to quantify cell movement.
2.8 Transwell cell invasion assay
Transwell chambers coated with Matrigel matrix (Corning, NY, USA) were utilized to perform Transwell cell invasion assays. THP-1 cells were seeded in the upper chamber with 200 μl serum-free RPMI-1640 medium at a density of 1 × 106 cells/ml. Then, 800 μl CM or fresh RPMI-1640 medium alone as a blank control for spontaneous invasion were added to the lower chamber. After incubation for 48 h, the numbers of invaded THP-1 cells within the lower chamber were photographed by Nikon Eclipse Ts2R inverted fluorescence microscope and counted by LUNA-II™ Automated Cell Counter.
2.9 Cell adhesion assay
Briefly, THP-1 cells were suspended in the CM or fresh RPMI-1640 medium alone as a blank control for spontaneous adhesion and seeded in the 24-well plate at the density of 2 × 104 cells/well. After 24 h, the wells were gently washed with PBS to remove non-adhered cells. The adherent THP-1 cells were then fixed with 4% PFA for 15 min and stained using DAPI (Beyotime, Shanghai, China) for 10 min at room temperature. Adherent cells were observed under Nikon Eclipse Ts2R inverted fluorescence microscope and counted using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.10 Macrophage differentiation and co-culture system
THP-1 monocytes were differentiated into macrophages as previously described [37]. In brief, the THP-1 cells were seeded into 6-well culture plates at 1 × 106 cells/ml in the presence of 100 ng/ml PMA for 24 h to obtain resting macrophages (M0). Subsequently, PMA was removed, and M0 cells were cultured with fresh RPMI-1640 medium for another 48 h to allow cellular recovery. To establish a co-culture system, the M0 cells were cultured in the CM of OSCC cell lines and harvested for further analysis after 72 h. CCL19 or CCL21 at 400 ng/ml was added into the FBS-free RPMI-1640 medium for 72 h to explore their effects on macrophage polarization in vitro.
2.11 Macrophage morphology and imaging
In order to investigate the effect of OSCC CM on macrophage morphology, co-cultured macrophages were fixed in 4% PFA and permeabilized by 0.2% Triton X-100 (Beyotime, Shanghai, China) beforehand. Actin-Tracker Red-555 Fluorescent Phalloidin (dilution 1:100, Beyotime, Shanghai, China) was applied to label the cytoskeleton, and DAPI (Beyotime, Shanghai, China) was employed to stain the nuclei. Fluorescently labelled cells were then observed and photographed under Nikon Eclipse Ts2R inverted fluorescence microscope.
2.12 Flow cytometry
Flow cytometry was performed to detect cytomembrane CD206 (also known as mannose receptor c-type 1, MRCI) expression of macrophage after co-cultured with CM. M0 macrophages without any further treatment were set up as the blank control group. Macrophages were washed, scraped off using cell scrapers and resuspended in ice-cold PBS containing 2% FBS. Human Fc-receptor block was used to reduce non-specific immunofluorescent staining. Next, cells were labelled with PE-conjugated mouse anti-human CD206 for 30 min at 4 °C in dark. At last, the proportion of CD206+ macrophages was analyzed by a BD FACSCanto II flow cytometer (BD Biosciences, San Jose, CA, USA) (10,000 events were recorded for each sample).
2.13 Immunofluorescence (IF) staining
Immunofluorescence (IF) staining was carried out to observe the CD163 expression level of CM-treated macrophages. After discarding culture medium, macrophages growing on coverslips were collected and washed with PBS containing 2% FBS for 3 times. Then, cells were fixed with 4% PFA at room temperature for 15 min and permeabilized by 0.2% Triton X-100 for 10 min. Unspecific binding sites were later blocked by QuickBlock™ Blocking Buffer (Beyotime, Shanghai, China) for 1 h at room temperature. The cells were incubated with the primary CD163 antibody conjugated to Alexa Fluor® 488 (dilution 1:200) at 4 °C in dark overnight. Then, samples were stained with DAPI for 10 min to label the nuclei. Finally, cells were observed and photographed by Nikon Eclipse Ts2R inverted fluorescence microscope.
2.14 Statistical analyses
All statistical analyses were conducted using GraphPad Prism 9 (GraphPad Software, Inc., CA, USA). The results were presented as the mean ± standard deviation (SD) of at least three different experiments. The significant difference between two groups was determined by independent sample or paired-sample Student's t test. The one-way analysis of variance (ANOVA) followed by the Dunnett's multiple comparisons test was employed to compare more than two groups. Spearman correlation analysis was applied to assess the statistical significance of correlations between two variables. Flow cytometry data analysis was performed using Flowjo software (Tree Star, Ashland, OR, USA). A two-tailed P < 0.05 was considered statistically significant if not especially noted.
3 Results
3.1 Overexpression of CCR7 positively correlated with lymph node metastasis and M2 macrophage infiltration in OSCC patients
In order to have a broad view of CCR7 expression pattern in pan-cancer scale, we downloaded RNAseq data of TCGA tumors from TIMER2.0 database. The results revealed that CCR7 was notably up-regulated in various tumor types, including HNSCC (red box) (Fig. 1A). Two GEO datasets were applied to further estimate CCR7 expression level in OSCC. We found that CCR7 was overexpressed in OSCC, whether viewed from the overall sample or the matched sample of a single patient (Fig. 1B). This generally in accordance with the immunohistochemical data obtained from HPA database, which showed that CCR7 was almost not expressed in normal oral epithelial tissue, while OSCC tissue had strong staining (Fig. 1C). Clinical information retrieved from TCGA-HNSC cohort was then employed to examine the clinicopathological significance of CCR7. As displayed in Fig. 1D, patients with severe lymph node involvement are more likely to overexpress CCR7 (P < 0.05).
CCR7 is overexpressed and positively correlated with M2 macrophage infiltration in OSCC patients. A CCR7 expression patterns across different tumour types in TIMER2.0. The red box represents HNSCC tissues and adjacent normal tissues. B CCR7 expression levels in OSCC tissues and normal oral tissues in GSE25099 and GSE74530 dataset. C CCR7 protein expression levels in OSCC tissues and normal oral epithelial tissues, determined by immunohistochemistry staining from HPA website. magnification, × 4. D Relationship between CCR7 expression and N stage of HNSCC patients, based on TCGA. E The association between CCR7 expression and the Immunescore of TCGA-HNSCC patients generated by ESTIMATE algorithm. F Correlation between CCR7 expression level and immune infiltration levels in TCGA-HNSCC patients based on TIMER2.0 database. G Correlation between CCR7 expression and M2 macrophage in TCGA-HNSCC patients, based on TIMER2.0 database. *P < 0.05, **P < 0.01, ***P < 0.001
Next, we investigated the latent role CCR7 plays in the tumor immunology of HNSCC. ESTIMATE algorithm demonstrated an obvious positive correlation between CCR7 and Immunescore in HNSCC patients (Fig. 1E, Spearman correlation coefficient = 0.7046, P < 0.001). Similarly, data from TIMER web server suggested that CCR7 had a noticeable effect on almost all types of immune cells in HNSCC (Fig. 1F). In view of the widely-discussed pro-tumor functions of M2 macrophages, we thus examined the association between CCR7 and M2 macrophages. Of note, 2 different algorithms (CIBERSOFT-ABS and QUANTISEQ) indicated that CCR7 considerably associated with M2 macrophage infiltration level in HNSCC (Fig. 1G; CIBERSOFT-ABS Spearman correlation coefficient = 0.581, P = 8.36e−46; QUANTISEQ Spearman correlation coefficient = 0.374, P = 8.89e−18). Meanwhile, spearman correlation analysis illustrated a strong correlation between CCR7 and two essential M2 macrophage biomarkers, CD163 and MRC1 (CD206), in HNSCC patients (Fig. 1G; CD163 Spearman correlation coefficient = 0.457, P = 9e−27; CD206 Spearman correlation coefficient = 0.346, P = 2.95e−15). The above results demonstrated that CCR7 is significantly up-regulated in OSCC patients and contributes to lymph node metastasis and M2 macrophage infiltration.
3.2 CCR7 deficiency in OSCC cells restrained macrophage recruitment and adhesion
To uncover the potential function of CCR7 regulating macrophage in OSCC, siCCR7 and siNC were utilized to transfect PCI-4B and PCI-37B cells. The results of qRT-PCR confirmed substantial CCR7 downregulation at mRNA levels 24 h post transfection (Fig. 2A). CM was collected as previously described from siNC transfected-OSCC cells (CM-siNC) and CCR7-silenced OSCC cells (CM-siCCR7), respectively. Then, we examined the migration and invasion abilities of macrophages after co-culture with CM from OSCC cells with different CCR7 expression levels. A sketch for the Transwell cell migration and invasion assay was supplied in Supplementary Fig. S1. Compared with blank control, we found that CM-siNC could significantly induce THP-1 cell migration. The CCR7 depletion, however, reverses this trend (Fig. 2B). Invasion assay exhibited a similar pattern (Fig. 2C). In line with our results, TCGA data pointed out that the mRNA expression level of CCR7 is positively related to CD68, a pan-marker of macrophages (Supplementary Fig. S2, Spearman correlation coefficient = 0.289, P = 6.27e−11). Moreover, THP-1 cells without any treatment barely attach to the dishes since they grow in suspension. We observed a dramatic increase of adherent THP-1 cells in the CM-siNC group while a reduction in the CM-siCCR7 treatment group. Collectively, these results indicated that CCR7 deficiency in OSCC cells confined macrophage accumulation and adhesion in the TME.
CCR7 deficiency in OSCC cells restrained macrophage recruitment and adhesion. A qRT-PCR examining CCR7 knockdown efficiency in OSCC cell lines PCI-4B and PCI-37B. B The effects of OSCC CM on the migration ability of THP-1 cells were quantified by Transwell cell migration assay. Representative images and statistical analysis histograms were shown (scale bar: 200 μm). C The effects of OSCC CM on the invasion ability of THP-1 cells were estimated by Transwell cell invasion assay. Representative images and statistical analysis histograms were shown (scale bar: 200 μm). D The effects of OSCC CM on the adhesion of THP-1 cells assessed by cell adhesion assay. The nuclei were labelled with DAPI (blue). Representative images and statistical analysis histograms were shown (scale bar: 200um). The results are presented as the mean ± SD, ns not significant, *P < 0.05, **P < 0.01, ***P < 0.001
3.3 Knockdown of CCR7 in OSCC cells suppressed macrophage M2-polarization
To further determine whether CCR7 was involved in the process of macrophage activation and polarization in OSCC, we induced suspension THP-1 monocytes into adherent M0 macrophages using PMA. The result of qRT-PCR showed that the mRNA level of pan macrophage marker CD68 was upregulated in M0 macrophages compared with THP-1 cells, suggesting a satisfying induction efficiency (Supplementary Fig. S3, P < 0.01). Subsequently, the M0 macrophages were cultivated with CM-siNC and CM-siCCR7 for 72 h, respectively. A sketch for the co-culture system was shown in Supplementary Fig. S4. Through the cytoskeleton staining, we observed that macrophages differ substantially in their morphologies when cultivating with CM with different CCR7 expression levels. Compared to untreated cells, macrophages tended to be larger and behaved more spindle and branched when treated with CM-siNC. Nevertheless, the macrophages were not only smaller but also appeared round and smooth in response to incubation with CM-siCCR7, similar to M0 macrophages (Fig. 3A). On the other hand, according to Fig. 3B, the mRNA levels of M2 macrophage markers CD163, CD206, interleukin-10 (IL-10) and transforming growth factor-β1 (TGF-β1) were upregulated in the CM-siNC groups compared with the untreated M0 group. By contrast, after treatment with CM-siCCR7, the mRNA levels of the above markers were markedly downregulated. We also employed flow cytometry to quantify the membrane expression of CD206. We noticed that nearly 20% of macrophages displayed positive staining of CD206 when coculturing with CM-siNC, whereas the frequency of CD206+ macrophages significantly decreased in CM-siCCR7 the group (Fig. 3C). Consistently, IF staining analysis revealed that the fluorescence intensity of CD163 was considerably higher in the CM-siNC group than in the blank control group and CM-siCCR7 group (Fig. 3D). Combined, these results presented evidence that knockdown of CCR7 in OSCC cells inhibited TAMs polarization to an M2 phenotype.
Knockdown of CCR7 in OSCC cells suppressed macrophage M2-polarization. A Representative morphological images of THP-1 derived macrophages cultivated with OSCC CM. The cytoskeleton was labelled with Actin-Tracker Red-555 Fluorescent Phalloidin (red), and nuclei were stained with DAPI (blue) (scale bar: 50 μm). B The mRNA expression level of CD163, CD206, IL-10 and TGF-β1 of macrophages co-cultured with OSCC CM, evaluated by qRT-PCR. C Representative Scatter plots showed the CD206 expression level of macrophages incubated with OSCC CM, measured by flow cytometry. The histogram displayed statistical analysis for the proportion of CD206+ macrophages. D Representative fluorescent images of macrophages treated with OSCC CM. The cytomembrane CD163 was labelled with Alexa Fluor® 488 (green), and nuclei were stained with DAPI (blue) (scale bar: 50um). The results are presented as the mean ± SD, ns: not significant, *P < 0.05, **P < 0.01, ***P < 0.001
3.4 CCL19 and CCL21 promoted macrophage M2-polarization in vitro
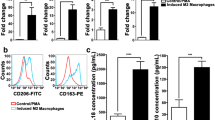
CCL19 and CCL21 are two primary functional ligands for CCR7 [38]. The results based on the TCGA database displayed a positively correlation between CCR7 and both CCL19 and CCL21 in OSCC (Fig. 4A, CCL19 Spearman correlation coefficient = 0.736, P = 5.23e−85; CCL21 Spearman correlation coefficient = 0.413, P = 1.02e−21). Previous scholars have proved that they can be massively secreted by OSCC cell lines PCI-4B and PCI-37B [39]. What is more, data from TIMER2.0 showed that both CCL19 and CCL21 correlated with macrophage M2-polarization (Fig. 4B, CCL19 Spearman correlation coefficient = 0.554, P = 7.40e−41; CCL21 Spearman correlation coefficient = 0.319, P = 4.39e−13). Hence, we supposed that CCL19 and CCL21 were involved in M2 polarization of macrophages. In vitro experiments were then carried out to identify the effects of recombinant human CCL19 and CCL21 on THP-1 derived macrophages. The results suggested that CCL19 and CCL21 (both at 400 ng/ml) enhanced mRNA expression level of various M2 markers in vitro (Fig. 4C). Also, the treatment of CCL19&21 significantly increased the proportion of CD206+ macrophages (Fig. 4D). Overall, current evidence seemed to support that CCL19 and CCL21 promoted M2-polarization of macrophages in vitro.
CCL19 and CCL21 promoted macrophage M2-polarization in vitro. A Spearman correlation analysis of CCR7 expression with CCL19 and CCL21, based on TIMER2.0 database. B Correlation between CCR7 expression and M2 macrophage, based on TIMER2.0 database. C The mRNA expression level of CD163, CD206, IL-10 and TGF-β1 of THP-1 derived macrophages stimulated by 400 ng/ml recombinant human CCL19 and CCL21, evaluated by qRT-PCR. D Representative Scatter plots showed the CD206 expression level of macrophages incubated with 400 ng/ml recombinant human CCL19 and CCL21, measured by flow cytometry. The histogram displayed statistical analysis for the proportion of CD206+ macrophages. The results are presented as the mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001
4 Discussion
Recent studies support the notion that cancer fate, including malignant progression, intravasation and therapy resistance, results not only from inner genetic changes of tumor cell itself but also from the complicated communication networks that it establishes with surrounding TME [40, 41]. The emerging treatment modality of immunotherapy has been developed, targeting TME to enhance the immune response to tumors and limit the growth of cancer cells [42]. The TME of OSCC is highly immunosuppressive and consists of many different subsets of cells [43], in which TAMs constitute a principal component [15]. It is considered that M2-like TAMs favor tumor growth via inducing tissue remodeling, suppressing adaptive immunity by T-cells, and producing a battery of bioactive molecules [44, 45]. Despite numerous studies regarding this essential topic, the molecular mechanism by which TAMs were recruited and activated is still far from clear. CCR7 is a G protein‐coupled transmembrane chemokine with multiple functions [46]. Our previous studies have verified that CCR7 is associated with the invasion and metastasis of tumors through eliciting a series of intracellular signaling pathways in OSCC [47, 48]. Nonetheless, little is known about the clinicopathological significance of CCR7 in TAMs infiltration. By establishing a co-culture system, our data support a hypothesis that OSCC cells actively recruit peripheral monocytes into TME and promote their polarization towards M2 macrophages through the overexpression of CCR7. Regulation of TAMs is an essential means in the immunotherapy of OSCC patients. Based on current results, depleting TAMs or suppressing their polarization from an M1 to an M2 phenotype by targeting CCR7 may be a novel therapeutic approach and deserves further clinical verification in real world study.
Previous studies postulated that CCR7 is abnormally upregulated in a number of malignancies such as breast cancer [49], lung adenocarcinoma [50] and gastric cancer [51]. In this work, we demonstrated that CCR7 was overexpressed in OSCC. The protein expression data obtained from HPA was generally consistent. Moreover, patients with advanced stages of OSCC are more likely to overexpress CCR7. Shang et al. [52] also discovered that CCR7 expression was significantly higher in OSCC patients with lymph node metastasis compared with those without (P = 0.015) and was also associated with tumor size (P = 0.014) and clinical stage (P = 0.009). These results indicated that CCR7 might serve as an oncogene in OSCC. Given that CCR7 is known to mediate many events in adaptive immune system [53], we next examined the relationship between CCR7 and immune infiltration level in OSCC. ESTIMATE algorithm and TIMER web server revealed that CCR7 expression level was positively associated with increased immune infiltration level in OSCC. Previous studies focused more on its distinct role on regulating T cells homing, activation and recruitment [54, 55]. The biological link between CCR7 and TAMs in OSCC is yet to be illustrated so far. The tumor-infiltrating immune cell models in TIMER2.0 database depicted that the expression of CCR7 promotes M2 macrophages infiltration level and correlates with M2 biomarkers in OSCC. Thereby, it is reasonable to speculate that CCR7 facilitate the malignant process of OSCC by modulating macrophage.
TAMs in the TME are mainly derived from peripheral blood monocytic precursors [44]. Therefore, recruitment of circulating monocytes is of vital importance in the inflammatory and immune responses of TME. Cytokines documented to have chemotactic effects for monocytes include CCL2 [56], CCL15 [57], CCL20 [58], vascular endothelial growth factor (VEGF) [59] and platelet-derived growth factor (PDGF) [60]. When treated with CM-siNC, we noticed an obvious increase in the migrated and invaded THP-1 cells, while this trend was reversed after silencing CCR7. Likewise, Yang et al. reported CCR7 is positively correlated with macrophage migration [61]. It is worth mentioning that CM-siNC enhanced adhesion of THP-1 cells compared to untreated cells, but this phenomenon was less seen in the CM-siCCR7 group. The THP-1 monocytic cell line grows in suspension and does not attach to cell culture surfaces in most cases. Adherence is considered a feature of its maturity, signaling that monocytes have started the differentiation into macrophages [62]. In other words, CCR7 contributed to TAMs accumulation in the TME of OSCC and may induce activation.
It is well acknowledged that macrophages are a rather dynamic cell population, exhibiting remarkable phenotypic heterogeneity and functional plasticity in response to exposure of TME stimuli [21]. We thus established a co-culture system to study the role CCR7 plays in the interaction between macrophages and OSCC cells. Our data suggested that CM-siNC activated macrophages to become M2-like phenotype, whereas a substantial decrease in the expression of M2 macrophage markers (CD163, CD206, IL-10, TGF-β1) was observed after co-cultured with CM-siCCR7. Results from flow cytometry showed that CM-siCCR7 reduced the proportion of CD206+ macrophages in vitro. IF staining data also implied that compared to untreated group, the fluorescence intensity of CD163 was dramatically higher in CM-siNC group, while it barely changed in CM-siCCR7 group. Sunil et al. yielded a similar result, who indicated that compared with wild-type, CCR7-knockout mice presented significantly lower levels of two well-acknowledged M2 macrophage markers (IL-10 and TGF-β1) [63]. In brief, these results reminded us that CCR7 might be an inducer of M2-like TAMs polarization in OSCC.
CCL19 and CCL21 are two natural ligands of CCR7 that widely exist in lymphoid organs and immune cells [46], and have been validated to be produced in large quantities by CCR7 high-expressed OSCC cell lines PCI-4B and PCI-37B [39]. Several recent studies have pointed to a strong association between CCL19&21 and cancer progression [64, 65]. In fact, earlier researchers have highlighted the indispensable roles CCL19 and CCL21 played in attracting TAMs. Cai et al. revealed that CCL19 and CCL21 treatment significantly increased the migration of monocytes, whereas the addition of CCR7-neutralizing antibody abolished both CCL19- and CCL21-induced monocyte migration [66]. Allaire et al. found that in monocytes, CCL19 binding to CCR7 results in potent phosphorylation of MAPK and leads to monocyte migration [67]. Similarly, Ato et al. reported that CCL19 and CCL21 stimulated macrophage migration in vitro in a dose-dependent fashion [68]. More importantly, results from TIMER2.0 database reminded us there is strong relationship between CCL19&21 and M2 macrophages in HNSCC. Accordingly, we conjectured they were involved in the M2 polarization of macrophages. As expected, recombinant human CCL19 and CCL21 largely reeducated M0 macrophage to M2 phenotype in vitro. Our results were corroborated by an in vivo study by Shields et al. [69]. They used short hairpin RNA (shRNA) to knockdown endogenous CCL21 secretion in murine melanoma cells and found that CCL21low tumors contained lower amounts of TGF-β1. TGF-β1 is not only a known M2 macrophage marker but also a critical immunomodulator that can shift the macrophage populations from M1 to pro-tumoral M2 phenotype [70]. Researchers also added CCL19 directly to cultures of human peripheral blood mononuclear cell (PBMC), and observed a significant enhancement of M2 marker IL-10 production [71]. In agreement with this, Pickens et al. demonstrated that CCL19 and CCL21 induce secretion of proangiogenic factors from macrophages [72], which is among the most crucial characteristics of M2 macrophages. Taken together, the present study implied that OSCC cells could educate TAMs toward M2 phenotype that promotes tumor growth and spread via, at least in part, CCR7-CCL19&21 axis.
CCL19&21 binding to CCR7 causes conformational changes, triggers various signaling cascades and elicit their biologic functions [73]. Our previous studies have uncovered that CCL19 could induce phosphorylation of JAK2/STAT3 and NF-κB, activate MAPK and PI3K/Akt signaling pathway through interaction with CCR7 [47, 48, 74, 75]. Similarly, Chen et al. [26] found that CCR7/CCL21 axis activated the JAK2/STAT3 signaling pathway. Many investigators have reported that the acquisition of TAM M2 phenotype is closely associated with pathways mentioned above, especially NF-κB [37, 76,77,78]. Hence, it is possible that the interaction of CCR7 and CCL19 or CCL21 promotes macrophage M2 polarization via one of these signaling pathways. Although the underlying mechanism is unable to be clarified by now, this will be the focus to investigate in our follow-up studies.
Nevertheless, it is undeniable that several potential limitations should be taken into consideration when interpreting our results. Firstly, since we focused on the biological functions of CCR7, two CCR7 high-expressed OSCC cell lines, PCI-4B and PCI-37B, were applied in this study. More types of tumor cell lines should be used in future analysis. Secondly, despite THP-1 cell line resembles native macrophages in morphological and functional properties and is commonly used as an appropriate model to imitate TAMs differentiation in vitro [79], some argue that heterogeneity still exists between THP-1 cells and primary cells [80]. Consequently, additional investigations using primary human macrophages are warranted to reach a more convincing conclusion. Last but not least, our findings should be further verified in a suitable immunocompetent OSCC animal model.
5 Conclusion
In conclusion, our data demonstrated that CCR7 was up-regulated in OSCC patients and correlated with increased M2 macrophage infiltration levels. Further in vitro investigation favored the assumption that CCR7 in OSCC cells promoted recruitment and M2-polarization of THP-1 derived macrophages, which was mainly modulated by CCL19 and CCL21. Although it requires further clinical verification, our work provided a potential CCR7-based antitumor strategy for OSCC patients.
Data availability
The datasets generated during and/or analyzed during the current study are available in the TIMER2.0 repository, http://timer.comp-genomics.org/; GEO repository, https://www.ncbi.nlm.nih.gov/geo/; HPA repository, https://www.proteinatlas.org/; TCGA repository, https://tcgadata.nci.nih.gov/tcga; ESTIMATE repository, https://sourceforge.net/projects/estimateproject/; TIMER repository, https://cistrome.shinyapps.io/timer/.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Shield KD, Ferlay J, Jemal A, Sankaranarayanan R, Chaturvedi AK, Bray F, Soerjomataram I. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J Clin. 2017;67(1):51–64. https://doi.org/10.3322/caac.21384.
Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma—an update. CA Cancer J Clin. 2015;65(5):401–21. https://doi.org/10.3322/caac.21293.
Miranda-Filho A, Bray F. Global patterns and trends in cancers of the lip, tongue and mouth. Oral Oncol. 2020;102: 104551. https://doi.org/10.1016/j.oraloncology.2019.104551.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. https://doi.org/10.3322/caac.21654.
Oliver AJ, Lau PKH, Unsworth AS, Loi S, Darcy PK, Kershaw MH, Slaney CY. Tissue-dependent tumor microenvironments and their impact on immunotherapy responses. Front Immunol. 2018;9:70. https://doi.org/10.3389/fimmu.2018.00070.
Farlow JL, Brenner JC, Lei YL, Chinn SB. Immune deserts in head and neck squamous cell carcinoma: a review of challenges and opportunities for modulating the tumor immune microenvironment. Oral Oncol. 2021;120: 105420. https://doi.org/10.1016/j.oraloncology.2021.105420.
Economopoulou P, Kotsantis I, Psyrri A. Tumor microenvironment and immunotherapy response in head and neck cancer. Cancers (Basel). 2020. https://doi.org/10.3390/cancers12113377.
Kujan O, Agag M, Smaga M, Vaishnaw Y, Idrees M, Shearston K, Farah CS. PD-1/PD-L1, Treg-related proteins, and tumour-infiltrating lymphocytes are associated with the development of oral squamous cell carcinoma. Pathology. 2022;54(4):409–16. https://doi.org/10.1016/j.pathol.2021.09.013.
Locati M, Curtale G, Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol. 2020;15:123–47. https://doi.org/10.1146/annurev-pathmechdis-012418-012718.
Ireland LV, Mielgo A. Macrophages and fibroblasts, key players in cancer chemoresistance. Front Cell Dev Biol. 2018;6:131. https://doi.org/10.3389/fcell.2018.00131.
Wenes M, Shang M, Di Matteo M, Goveia J, Martín-Pérez R, Serneels J, Prenen H, Ghesquière B, Carmeliet P, Mazzone M. Macrophage metabolism controls tumor blood vessel morphogenesis and metastasis. Cell Metab. 2016;24(5):701–15. https://doi.org/10.1016/j.cmet.2016.09.008.
Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12(1):76. https://doi.org/10.1186/s13045-019-0760-3.
Yang Q, Guo N, Zhou Y, Chen J, Wei Q, Han M. The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm Sin B. 2020;10(11):2156–70. https://doi.org/10.1016/j.apsb.2020.04.004.
Costa NL, Valadares MC, Souza PP, Mendonça EF, Oliveira JC, Silva TA, Batista AC. Tumor-associated macrophages and the profile of inflammatory cytokines in oral squamous cell carcinoma. Oral Oncol. 2013;49(3):216–23. https://doi.org/10.1016/j.oraloncology.2012.09.012.
Alves AM, Diel LF, Lamers ML. Macrophages and prognosis of oral squamous cell carcinoma: a systematic review. J Oral Pathol Med. 2018;47(5):460–7. https://doi.org/10.1111/jop.12643.
Huang Z, Xie N, Liu H, Wan Y, Zhu Y, Zhang M, Tao Y, Zhou H, Liu X, Hou J, et al. The prognostic role of tumour-infiltrating lymphocytes in oral squamous cell carcinoma: a meta-analysis. J Oral Pathol Med. 2019;48(9):788–98. https://doi.org/10.1111/jop.12927.
Hadler-Olsen E, Wirsing AM. Tissue-infiltrating immune cells as prognostic markers in oral squamous cell carcinoma: a systematic review and meta-analysis. Br J Cancer. 2019;120(7):714–27. https://doi.org/10.1038/s41416-019-0409-6.
Brown JM, Recht L, Strober S. The promise of targeting macrophages in cancer therapy. Clin Cancer Res. 2017;23(13):3241–50. https://doi.org/10.1158/1078-0432.Ccr-16-3122.
Porcheray F, Viaud S, Rimaniol AC, Léone C, Samah B, Dereuddre-Bosquet N, Dormont D, Gras G. Macrophage activation switching: an asset for the resolution of inflammation. Clin Exp Immunol. 2005;142(3):481–9. https://doi.org/10.1111/j.1365-2249.2005.02934.x.
Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci. 2021. https://doi.org/10.3390/ijms22136995.
Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. https://doi.org/10.1146/annurev-immunol-032713-120145.
Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559–72. https://doi.org/10.1038/nri.2017.49.
Bill CA, Allen CM, Vines CM. C-C chemokine receptor 7 in cancer. Cells. 2022. https://doi.org/10.3390/cells11040656.
Xu Z, Zheng X, Yang L, Liu F, Zhang E, Duan W, Bai S, Safdar J, Li Z, Sun C. Chemokine receptor 7 promotes tumor migration and invasiveness via the RhoA/ROCK pathway in metastatic squamous cell carcinoma of the head and neck. Oncol Rep. 2015;33(2):849–55. https://doi.org/10.3892/or.2014.3631.
Chen Y, Shao Z, Jiang E, Zhou X, Wang L, Wang H, Luo X, Chen Q, Liu K, Shang Z. CCL21/CCR7 interaction promotes EMT and enhances the stemness of OSCC via a JAK2/STAT3 signaling pathway. J Cell Physiol. 2020;235(9):5995–6009. https://doi.org/10.1002/jcp.29525.
Liu MD, Wu H, Wang S, Pang P, Jin S, Sun CF, Liu FY. MiR-1275 promotes cell migration, invasion and proliferation in squamous cell carcinoma of head and neck via up-regulating IGF-1R and CCR7. Gene. 2018;646:1–7. https://doi.org/10.1016/j.gene.2017.12.049.
Wang S, Jin S, Liu MD, Pang P, Wu H, Qi ZZ, Liu FY, Sun CF. Hsa-let-7e-5p inhibits the proliferation and metastasis of head and neck squamous cell carcinoma cells by targeting chemokine receptor 7. J Cancer. 2019;10(8):1941–8. https://doi.org/10.7150/jca.29536.
Zhang Z, Liu F, Li Z, Wang D, Li R, Sun C. Jak3 is involved in CCR7-dependent migration and invasion in metastatic squamous cell carcinoma of the head and neck. Oncol Lett. 2017;13(5):3191–7. https://doi.org/10.3892/ol.2017.5861.
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–14. https://doi.org/10.1093/nar/gkaa407.
Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. https://doi.org/10.1093/nar/30.1.207.
Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017. https://doi.org/10.1126/science.aan2507.
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. https://doi.org/10.1038/ncomms3612.
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–10. https://doi.org/10.1158/0008-5472.Can-17-0307.
Wang J, Xi L, Hunt JL, Gooding W, Whiteside TL, Chen Z, Godfrey TE, Ferris RL. Expression pattern of chemokine receptor 6 (CCR6) and CCR7 in squamous cell carcinoma of the head and neck identifies a novel metastatic phenotype. Cancer Res. 2004;64(5):1861–6. https://doi.org/10.1158/0008-5472.can-03-2968.
Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, Ferris RL, Lai SY. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29(2):163–88. https://doi.org/10.1002/hed.20478.
Dan H, Liu S, Liu J, Liu D, Yin F, Wei Z, Wang J, Zhou Y, Jiang L, Ji N, et al. RACK1 promotes cancer progression by increasing the M2/M1 macrophage ratio via the NF-κB pathway in oral squamous cell carcinoma. Mol Oncol. 2020;14(4):795–807. https://doi.org/10.1002/1878-0261.12644.
Salem A, Alotaibi M, Mroueh R, Basheer HA, Afarinkia K. CCR7 as a therapeutic target in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875(1): 188499. https://doi.org/10.1016/j.bbcan.2020.188499.
Wang J, Seethala RR, Zhang Q, Gooding W, van Waes C, Hasegawa H, Ferris RL. Autocrine and paracrine chemokine receptor 7 activation in head and neck cancer: implications for therapy. J Natl Cancer Inst. 2008;100(7):502–12. https://doi.org/10.1093/jnci/djn059.
Rangel R, Pickering CR, Sikora AG, Spiotto MT. Genetic changes driving immunosuppressive microenvironments in oral premalignancy. Front Immunol. 2022;13: 840923. https://doi.org/10.3389/fimmu.2022.840923.
Elmusrati A, Wang J, Wang CY. Tumor microenvironment and immune evasion in head and neck squamous cell carcinoma. Int J Oral Sci. 2021;13(1):24. https://doi.org/10.1038/s41368-021-00131-7.
Kujan O, van Schaijik B, Farah CS. Immune checkpoint inhibitors in oral cavity squamous cell carcinoma and oral potentially malignant disorders: a systematic review. Cancers (Basel). 2020. https://doi.org/10.3390/cancers12071937.
Chen SMY, Krinsky AL, Woolaver RA, Wang X, Chen Z, Wang JH. Tumor immune microenvironment in head and neck cancers. Mol Carcinog. 2020;59(7):766–74. https://doi.org/10.1002/mc.23162.
Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. https://doi.org/10.1016/j.immuni.2014.06.010.
Petruzzi MN, Cherubini K, Salum FG, de Figueiredo MA. Role of tumour-associated macrophages in oral squamous cells carcinoma progression: an update on current knowledge. Diagn Pathol. 2017;12(1):32. https://doi.org/10.1186/s13000-017-0623-6.
Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8(5):362–71. https://doi.org/10.1038/nri2297.
Liu FY, Safdar J, Li ZN, Fang QG, Zhang X, Xu ZF, Sun CF. CCR7 regulates cell migration and invasion through MAPKs in metastatic squamous cell carcinoma of head and neck. Int J Oncol. 2014;45(6):2502–10. https://doi.org/10.3892/ijo.2014.2674.
Liu FY, Safdar J, Li ZN, Fang QG, Zhang X, Xu ZF, Sun CF. CCR7 regulates cell migration and invasion through JAK2/STAT3 in metastatic squamous cell carcinoma of the head and neck. Biomed Res Int. 2014;2014: 415375. https://doi.org/10.1155/2014/415375.
Mohammed MM, Shaker O, Ramzy MM, Gaber SS, Kamel HS, Abed El Baky MF. The relation between ACKR4 and CCR7 genes expression and breast cancer metastasis. Life Sci. 2021;279: 119691. https://doi.org/10.1016/j.lfs.2021.119691.
Liu F, Wu H. CC chemokine receptors in lung adenocarcinoma: the inflammation-related prognostic biomarkers and immunotherapeutic targets. J Inflamm Res. 2021;14:267–85. https://doi.org/10.2147/jir.S278395.
Zhou R, Sun J, He C, Huang C, Yu H. CCL19 suppresses gastric cancer cell proliferation, migration, and invasion through the CCL19/CCR7/AIM2 pathway. Hum Cell. 2020;33(4):1120–32. https://doi.org/10.1007/s13577-020-00375-1.
Shang ZJ, Liu K, Shao Z. Expression of chemokine receptor CCR7 is associated with cervical lymph node metastasis of oral squamous cell carcinoma. Oral Oncol. 2009;45(6):480–5. https://doi.org/10.1016/j.oraloncology.2008.06.005.
Comerford I, Harata-Lee Y, Bunting MD, Gregor C, Kara EE, McColl SR. A myriad of functions and complex regulation of the CCR7/CCL19/CCL21 chemokine axis in the adaptive immune system. Cytokine Growth Factor Rev. 2013;24(3):269–83. https://doi.org/10.1016/j.cytogfr.2013.03.001.
Moschovakis GL, Förster R. Multifaceted activities of CCR7 regulate T-cell homeostasis in health and disease. Eur J Immunol. 2012;42(8):1949–55. https://doi.org/10.1002/eji.201242614.
Choi H, Song H, Jung YW. the Roles of CCR7 for the homing of memory CD8+ T cells into their survival niches. Immune Netw. 2020;20(3): e20. https://doi.org/10.4110/in.2020.20.e20.
Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–5. https://doi.org/10.1038/nature10138.
Liu LZ, Zhang Z, Zheng BH, Shi Y, Duan M, Ma LJ, Wang ZC, Dong LQ, Dong PP, Shi JY, et al. CCL15 recruits suppressive monocytes to facilitate immune escape and disease progression in hepatocellular carcinoma. Hepatology. 2019;69(1):143–59. https://doi.org/10.1002/hep.30134.
Nandi B, Shapiro M, Samur MK, Pai C, Frank NY, Yoon C, Prabhala RH, Munshi NC, Gold JS. Stromal CCR6 drives tumor growth in a murine transplantable colon cancer through recruitment of tumor-promoting macrophages. Oncoimmunology. 2016;5(8): e1189052. https://doi.org/10.1080/2162402x.2016.1189052.
Huang D, Song SJ, Wu ZZ, Wu W, Cui XY, Chen JN, Zeng MS, Su SC. Epstein-Barr virus-induced VEGF and GM-CSF drive nasopharyngeal carcinoma metastasis via recruitment and activation of macrophages. Cancer Res. 2017;77(13):3591–604. https://doi.org/10.1158/0008-5472.Can-16-2706.
Yang Y, Andersson P, Hosaka K, Zhang Y, Cao R, Iwamoto H, Yang X, Nakamura M, Wang J, Zhuang R, et al. The PDGF-BB-SOX7 axis-modulated IL-33 in pericytes and stromal cells promotes metastasis through tumour-associated macrophages. Nat Commun. 2016;7:11385. https://doi.org/10.1038/ncomms11385.
Yang X, Zhang J, Chen L, Yuan Z, Qin X, Wu Q, Shen D, He H, Yu C. The role of UNC5b in ox-LDL inhibiting migration of RAW264.7 macrophages and the involvement of CCR7. Biochem Biophys Res Commun. 2018;505(3):637–43. https://doi.org/10.1016/j.bbrc.2018.09.178.
Chimal-Ramírez GK, Espinoza-Sánchez NA, Chávez-Sánchez L, Arriaga-Pizano L, Fuentes-Pananá EM. Monocyte differentiation towards protumor activity does not correlate with M1 or M2 phenotypes. J Immunol Res. 2016;2016:6031486. https://doi.org/10.1155/2016/6031486.
Chauhan SK, Saban DR, Dohlman TH, Dana R. CCL-21 conditioned regulatory T cells induce allotolerance through enhanced homing to lymphoid tissue. J Immunol. 2014;192(2):817–23. https://doi.org/10.4049/jimmunol.1203469.
Gowhari Shabgah A, Jaber Al-Obaidi ZM, Rahman HS, Abdelbasset WK, Suksatan W, Bokov DO, Thangavelu L, Turki Jalil A, Jadidi-Niaragh F, Mohammadi H, et al. Does CCL19 act as a double-edged sword in cancer development? Clin Exp Immunol. 2021. https://doi.org/10.1093/cei/uxab039.
Rizeq B, Malki MI. The role of CCL21/CCR7 chemokine axis in breast cancer progression. Cancers (Basel). 2020. https://doi.org/10.3390/cancers12041036.
Cai W, Tao J, Zhang X, Tian X, Liu T, Feng X, Bai J, Yan C, Han Y. Contribution of homeostatic chemokines CCL19 and CCL21 and their receptor CCR7 to coronary artery disease. Arterioscler Thromb Vasc Biol. 2014;34(9):1933–41. https://doi.org/10.1161/atvbaha.113.303081.
Allaire MA, Dumais N. Involvement of the MAPK and RhoA/ROCK pathways in PGE2-mediated CCR7-dependent monocyte migration. Immunol Lett. 2012;146(1–2):70–3. https://doi.org/10.1016/j.imlet.2012.05.002.
Ato M, Nakano H, Kakiuchi T, Kaye PM. Localization of marginal zone macrophages is regulated by C-C chemokine ligands 21/19. J Immunol. 2004;173(8):4815–20. https://doi.org/10.4049/jimmunol.173.8.4815.
Shields JD, Kourtis IC, Tomei AA, Roberts JM, Swartz MA. Induction of lymphoidlike stroma and immune escape by tumors that express the chemokine CCL21. Science. 2010;328(5979):749–52. https://doi.org/10.1126/science.1185837.
Caja L, Dituri F, Mancarella S, Caballero-Diaz D, Moustakas A, Giannelli G, Fabregat I. TGF-β and the tissue microenvironment: relevance in fibrosis and cancer. Int J Mol Sci. 2018. https://doi.org/10.3390/ijms19051294.
Byrnes HD, Kaminski H, Mirza A, Deno G, Lundell D, Fine JS. Macrophage inflammatory protein-3 beta enhances IL-10 production by activated human peripheral blood monocytes and T cells. J Immunol. 1999;163(9):4715–20.
Pickens SR, Chamberlain ND, Volin MV, Pope RM, Mandelin AM 2nd, Shahrara S. Characterization of CCL19 and CCL21 in rheumatoid arthritis. Arthritis Rheum. 2011;63(4):914–22. https://doi.org/10.1002/art.30232.
Hauser MA, Legler DF. Common and biased signaling pathways of the chemokine receptor CCR7 elicited by its ligands CCL19 and CCL21 in leukocytes. J Leukoc Biol. 2016;99(6):869–82. https://doi.org/10.1189/jlb.2MR0815-380R.
Liu FY, Zhao ZJ, Li P, Ding X, Guo N, Yang LL, Zong ZH, Sun CF. NF-κB participates in chemokine receptor 7-mediated cell survival in metastatic squamous cell carcinoma of the head and neck. Oncol Rep. 2011;25(2):383–91. https://doi.org/10.3892/or.2010.1090.
Liu FY, Zhao ZJ, Li P, Ding X, Zong ZH, Sun CF. Mammalian target of rapamycin (mTOR) is involved in the survival of cells mediated by chemokine receptor 7 through PI3K/Akt in metastatic squamous cell carcinoma of the head and neck. Br J Oral Maxillofac Surg. 2010;48(4):291–6. https://doi.org/10.1016/j.bjoms.2009.06.007.
Zhang H, Zhao B, Wei H, Zeng H, Sheng D, Zhang Y. Cucurbitacin B controls M2 macrophage polarization to suppresses metastasis via targeting JAK-2/STAT3 signalling pathway in colorectal cancer. J Ethnopharmacol. 2022;287: 114915. https://doi.org/10.1016/j.jep.2021.114915.
Kawano A, Ariyoshi W, Yoshioka Y, Hikiji H, Nishihara T, Okinaga T. Docosahexaenoic acid enhances M2 macrophage polarization via the p38 signaling pathway and autophagy. J Cell Biochem. 2019;120(8):12604–17. https://doi.org/10.1002/jcb.28527.
Lian G, Chen S, Ouyang M, Li F, Chen L, Yang J. Colon cancer cell secretes EGF to promote M2 polarization of TAM through EGFR/PI3K/AKT/mTOR pathway. Technol Cancer Res Treat. 2019;18:1533033819849068. https://doi.org/10.1177/1533033819849068.
Chanput W, Mes JJ, Wichers HJ. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol. 2014;23(1):37–45. https://doi.org/10.1016/j.intimp.2014.08.002.
Tarique AA, Logan J, Thomas E, Holt PG, Sly PD, Fantino E. Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages. Am J Respir Cell Mol Biol. 2015;53(5):676–88. https://doi.org/10.1165/rcmb.2015-0012OC.
Acknowledgements
We would like to express our deepest and sincerest gratitude to Dr. Qi-Jun Wan and Mrs. Ji-Tao Zhou. It is their unconditional help and consistent support that make this work possible.
Funding
This study was supported by Liaoning Department of Education Research Foundation (Grant No. JC2019025), Liaoning Science and Technology Program (Grant No. 2019-ZD-0751) and Special Funds of “First-Class Universities and Disciplines of the World” Project (Grant No. 115-3110210730).
Author information
Authors and Affiliations
Contributions
Conceptualization: W-HZ, YW and C-FS; Methodology: W-HZ, CY, W-DD, MAA-A and F-YL; Formal analysis and investigation: W-HZ, YW, W-DD, LZ, S-FL, J-XG, SJ and Z-XW; Writing—original draft preparation: W-HZ; Writing—review and editing: YW, CY, W-DD, MAA-A, LZ, S-FL, J-XG, SJ, Z-XW, F-YL and C-FS; Funding acquisition: F-YL; Resources: F-YL and C-FS; Supervision: F-YL and C-FS. All authors have read and approved the final manuscript, and be willing to take responsibility for the entire manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All methods in this study were carried out in accordance with relevant guidelines and regulations. All human data in current study were retrieved from open public databases, which can be downloaded freely by users for scientific research and publication. No ethical issue was involved. Hence, this study was exempted from review by The Ethics Committee of School of Stomatology, China Medical University.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, WH., Wang, Y., Yan, C. et al. CC chemokine receptor 7 promotes macrophage recruitment and induces M2-polarization through CC chemokine ligand 19&21 in oral squamous cell carcinoma. Discov Onc 13, 67 (2022). https://doi.org/10.1007/s12672-022-00533-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-022-00533-x