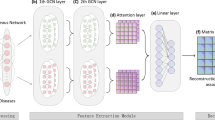
Abstract
Drug–drug interaction refers to taking the two drugs may produce certain reaction which may be a threat to patients’ health, or enhance the efficacy helpful for medical work. Therefore, it is necessary to study and predict it. In fact, traditional experimental methods can be used for drug–drug interaction prediction, but they are time-consuming and costly, so we prefer to use more accurate and convenient calculation methods to predict the unknown drug–drug interaction. In this paper, we proposed a deep learning framework called MSResG that considers multi-sources features of drugs and combines them with Graph Auto-Encoder to predicting. Firstly, the model obtains four feature representations of drugs from the database, namely, chemical substructure, target, pathway and enzyme, and then calculates the Jaccard similarity of the drugs. To balance different drug features, we perform similarity integration by finding the mean value. Then we will be comprehensive similarity network combined with drug interaction network, and encodes and decodes it using the graph auto-encoder based on residual graph convolution network. Encoding is to learn the potential feature vectors of drugs, which contain similar information and interaction information. Decoding is to reconstruct the network to predict unknown drug-drug interaction. The experimental results show that our model has advanced performance and is superior to other existing advanced methods. Case study also shows that MSResG has practical significance.
Graphical Abstract











Similar content being viewed by others
References
Foucquier J, Guedj M (2015) Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect 3:e00149. https://doi.org/10.1002/prp2.149
Kusuhara H (2014) How far should we go? Perspective of drug-drug interaction studies in drug development. Drug Metab Pharmacokinet 29:227–228. https://doi.org/10.2133/dmpk.DMPK-14-PF-903
Wishart DS, Feunang YD, Guo AC et al (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074–D1082. https://doi.org/10.1093/nar/gkx1037
Onakpoya IJ, Heneghan CJ, Aronson JK (2016) Post-marketing withdrawal of 462 medicinal products because of adverse drug reactions: a systematic review of the world literature. BMC Med 14:10. https://doi.org/10.1186/s12916-016-0553-2
Qiu Y, Zhang Y, Deng Y et al (2022) A comprehensive review of computational methods for drug-drug interaction detection. IEEE/ACM Trans Comput Biol Bioinf 19:1968–1985. https://doi.org/10.1109/TCBB.2021.3081268
He H, Chen G, Yu-Chian Chen C (2022) 3DGT-DDI: 3D graph and text based neural network for drug-drug interaction prediction. Brief Bioinform 23:bbac134. https://doi.org/10.1093/bib/bbac134
Yan C, Duan G, Zhang Y et al (2022) Predicting drug-drug interactions based on integrated similarity and semi-supervised learning. IEEE/ACM Trans Comput Biol Bioinform 19:168–179. https://doi.org/10.1109/TCBB.2020.2988018
Bag S, Kumar SK, Tiwari MK (2019) An efficient recommendation generation using relevant Jaccard similarity. Inf Sci 483:53–64. https://doi.org/10.1016/j.ins.2019.01.023
Xia P, Zhang L, Li F (2015) Learning similarity with cosine similarity ensemble. Inf Sci 307:39–52. https://doi.org/10.1016/j.ins.2015.02.024
Cheng F, Zhao Z (2014) Machine learning-based prediction of drug–drug interactions by integrating drug phenotypic, therapeutic, chemical, and genomic properties. J Am Med Inform Assoc 21:e278–e286. https://doi.org/10.1136/amiajnl-2013-002512
Shelhamer E, Long J, Darrell T (2017) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 39:640–651. https://doi.org/10.1109/CVPR.2015.7298965
Lipton ZC, Berkowitz J, Elkan C (2015) A critical review of recurrent neural networks for sequence learning. https://doi.org/10.48550/arXiv.1506.00019
Liu J, Lei X, Zhang Y, Pan Y (2023) The prediction of molecular toxicity based on BiGRU and GraphSAGE. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2022.106524
Liu X, Yang M (2022) Research on conversational machine reading comprehension based on dynamic graph neural network. J Integr Technol 11:67–78. https://doi.org/10.12146/j.issn.2095-3135.20211122001
Ryu JY, Kim HU, Lee SY (2018) Deep learning improves prediction of drug-drug and drug-food interactions. Proc Natl Acad Sci USA 115:E4304–E4311. https://doi.org/10.1073/pnas.1803294115
Feng Y-H, Zhang S-W, Shi J-Y (2020) DPDDI: a deep predictor for drug-drug interactions. BMC Bioinform 21:419. https://doi.org/10.1186/s12859-020-03724-x
Lin X, Quan Z, Wang Z-J, et al (2020) KGNN: knowledge graph neural network for drug-drug interaction prediction. In: Twenty-Ninth International Joint Conference on Artificial Intelligence, pp 2739–2745. https://doi.org/10.24963/ijcai.2020/380
Han X, Xie R, Li X, Li J (2022) SmileGNN: drug-drug interaction prediction based on the SMILES and graph neural network. Life (Basel) 12:319. https://doi.org/10.3390/life12020319
Li G, Müller M, Thabet A, Ghanem B (2019) DeepGCNs: can GCNs go as deep as CNNs? International Conference on Computer Vision 2019. https://doi.org/10.48550/arXiv.1904.03751
Wang F, Lei X, Liao B, Wu F-X (2022) Predicting drug-drug interactions by graph convolutional network with multi-kernel. Brief Bioinform 23:bbab511. https://doi.org/10.1093/bib/bbab511
Kipf TN, Welling M (2016) Variational graph auto-encoders. In: Conference and Workshop on Neural Information Processing Systems. https://doi.org/10.48550/arXiv.1611.07308
Zhang W, Chen Y, Liu F et al (2017) Predicting potential drug-drug interactions by integrating chemical, biological, phenotypic and network data. BMC Bioinform 18:18. https://doi.org/10.1186/s12859-016-1415-9
Wan F, Hong L, Xiao A et al (2019) NeoDTI: neural integration of neighbor information from a heterogeneous network for discovering new drug-target interactions. Bioinformatics 35:104–111. https://doi.org/10.1093/bioinformatics/bty543
Xie J, Zhao C, Ouyang J et al (2022) TP-DDI: a two-pathway deep neural network for drug-drug interaction prediction. Interdiscip Sci 14:895–905. https://doi.org/10.1007/s12539-022-00524-0
Schwarz K, Allam A, Perez Gonzalez NA, Krauthammer M (2021) AttentionDDI: Siamese attention-based deep learning method for drug-drug interaction predictions. BMC Bioinform 22:412. https://doi.org/10.1186/s12859-021-04325-y
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucl Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Kim S, Thiessen PA, Bolton EE et al (2016) PubChem substance and compound databases. Nucl Acids Res 44:D1202-1213. https://doi.org/10.1093/nar/gkv951
Lei S, Lei X, Liu L (2022) Drug repositioning based on heterogeneous networks and variational graph autoencoders. Front Pharmacol 13:5431. https://doi.org/10.3389/fphar.2022.1056605
Zhang Y, Lei X, Pan Y, Wu F-X (2022) Drug repositioning with GraphSAGE and clustering constraints based on drug and disease networks. Front Pharmacol 13:872785. https://doi.org/10.3389/fphar.2022.872785
Wang F, Ding Y, Lei X et al (2021) Human protein complex-based drug signatures for personalized cancer medicine. IEEE J Biomed Health Inform 25:4079–4088. https://doi.org/10.1109/JBHI.2021.3120933
Lahitani AR, Permanasari AE, Setiawan NA (2016) Cosine similarity to determine similarity measure: study case in online essay assessment. In: 2016 4th International Conference on Cyber and IT Service Management, 1–6. https://doi.org/10.1109/CITSM.2016.7577578
Kipf TN, Welling M (2017) Semi-supervised classification with graph convolutional networks. https://doi.org/10.48550/arXiv.1609.02907
An J, Guo L, Liu W et al (2021) IGAGCN: information geometry and attention-based spatiotemporal graph convolutional networks for traffic flow prediction. Neural Netw 143:355–367. https://doi.org/10.1016/j.neunet.2021.05.035
Zhu Y, Ma J, Yuan C, Zhu X (2022) Interpretable learning based dynamic graph convolutional networks for Alzheimer’s disease analysis. Inform Fus 77:53–61. https://doi.org/10.1016/j.inffus.2021.07.013
Chipofya M, Tayara H, Chong KT (2021) Drug therapeutic-use class prediction and repurposing using graph convolutional networks. Pharmaceutics 13:1906. https://doi.org/10.3390/pharmaceutics13111906
Ding Y, Lei X, Liao B, Wu F-X (2022) Predicting miRNA-disease associations based on multi-view variational graph auto-encoder with matrix factorization. IEEE J Biomed Health Inform 26:446–457. https://doi.org/10.1109/JBHI.2021.3088342
Zhang T, Gu J, Wang Z et al (2022) Protein subcellular localization prediction model based on graph convolutional network. Interdiscip Sci Comput Life Sci 14:937–946. https://doi.org/10.1007/s12539-022-00529-9
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. IEEE Conf Comput Vis Pattern Recogn (CVPR) 2016:770–778. https://doi.org/10.1109/CVPR.2016.90
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. https://doi.org/10.48550/arXiv.1412.6980
Srivastava N, Hinton G, Krizhevsky A, et al (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15:1929–1958. http://jmlr.org/papers/v15/srivastava14a.html
Smith LN (2017) Cyclical learning rates for training neural networks. In: 2017 IEEE Winter Conference on Applications of Computer Vision (WACV) 464–472. https://doi.org/10.48550/arXiv.1506.01186
Chiang W-L, Liu X, Si S, et al (2019) Cluster-GCN: an efficient algorithm for training deep and large graph convolutional networks. In: Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 257–266. https://doi.org/10.48550/arXiv.1905.07953
Pei Y, Huang T, Ipenburg W, Pechenizkiy M (2022) ResGCN: attention-based deep residual modeling for anomaly detection on attributed networks. Mach Learn 111:1–23. https://doi.org/10.1109/DSAA53316.2021.9564233
Deng Y, Xu X, Qiu Y et al (2020) A multimodal deep learning framework for predicting drug–drug interaction events. Bioinformatics 36:4316–4322. https://doi.org/10.1093/bioinformatics/btaa501
Zhang P, Wang F, Hu J, Sorrentino R (2015) Label propagation prediction of drug-drug interactions based on clinical side effects. Sci Rep 5:12339. https://doi.org/10.1038/srep12339
Abeywickrama T, Cheema M, Taniar D (2016) k-nearest neighbors on road networks: a journey in experimentation and in-memory implementation. Proc VLDB Endowm 9:492–503. https://doi.org/10.14778/2904121.2904125
Lü L, Pan L, Zhou T et al (2015) Toward link predictability of complex networks. Proc Natl Acad Sci USA 112:2325–2330. https://doi.org/10.1073/pnas.1424644112
Vilar S, Harpaz R, Uriarte E et al (2012) Drug-drug interaction through molecular structure similarity analysis. J Am Med Inform Assoc 19:1066–1074. https://doi.org/10.1136/amiajnl-2012-000935
Vilar S, Uriarte E, Santana L, Tatonetti N (2013) Detection of drug-drug interactions by modeling interaction profile fingerprints. PLoS ONE 8:e58321. https://doi.org/10.1371/journal.pone.0058321
Davis AP, Grondin CJ, Johnson RJ et al (2021) Comparative toxicogenomics database (CTD): update 2021. Nucl Acids Res 49:D1138–D1143. https://doi.org/10.1093/nar/gkaa891
Funding
This work was supported by the National Natural Science Foundation of China under Grants 62272288, 61972451, 61902230 and U22A2041, and the Shenzhen Science and Technology Program under Grant KQTD20200820113106007.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, L., Lei, X., Chen, M. et al. MSResG: Using GAE and Residual GCN to Predict Drug–Drug Interactions Based on Multi-source Drug Features. Interdiscip Sci Comput Life Sci 15, 171–188 (2023). https://doi.org/10.1007/s12539-023-00550-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-023-00550-6