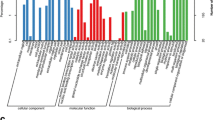
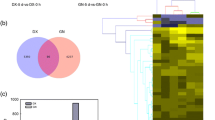
Abstract
Ziziphus nummularia an elite heat-stress tolerant shrub, grows in arid regions of desert. However, its molecular mechanism responsible for heat stress tolerance is unexplored. Therefore, we analysed whole transcriptome of Jaisalmer (heat tolerant) and Godhra (heat sensitive) genotypes of Z. nummularia to understand its molecular mechanism responsible for heat stress tolerance. De novo assembly of 16,22,25,052 clean reads yielded 276,029 transcripts. A total of 208,506 unigenes were identified which contains 4290 and 1043 differentially expressed genes (DEG) in TGO (treated Godhra at 42 °C) vs. CGO (control Godhra) and TJR (treated Jaisalmer at 42 °C) vs. CJR (control Jaisalmer), respectively. A total of 987 (67 highly enriched) and 754 (34 highly enriched) pathways were obsorved in CGO vs. TGO and CJR vs. TJR, respectively. Antioxidant pathways and TFs like Homeobox, HBP, ARR, PHD, GRAS, CPP, and E2FA were uniquely observed in Godhra genotype and SET domains were uniquely observed in Jaisalmer genotype. Further transposable elements were highly up-regulated in Godhra genotype but no activation in Jaisalmer genotype. A total of 43,093 and 39,278 simple sequence repeats were identified in the Godhra and Jaisalmer genotypes, respectively. A total of 10 DEGs linked to heat stress were validated in both genotypes for their expression under different heat stresses using quantitative real-time PCR. Comparing expression patterns of the selected DEGs identified ClpB1 as a potential candidate gene for heat tolerance in Z. nummularia. Here we present first characterized transcriptome of Z. nummularia in response to heat stress for the identification and characterization of heat stress-responsive genes.







Similar content being viewed by others
References
Abbas F, Rehman I, Adrees M, Ibrahim M, Saleem F, Ali S, Rizwan M, Salik MR (2018) Prevailing trends of climatic extremes across Indus-Delta of Sindh-Pakistan. Theor Appl Climatol 131(3–4):1101–1117. https://doi.org/10.1007/s00704-016-2028-y
Agarwal PK, Jha B (2010) Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol Plant 54:201–212. https://doi.org/10.1007/s10535-010-0038-7
Ambawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 19(3):307–321. https://doi.org/10.1007/s12298-013-0179-1
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Chakraborty K, Sairam RK, Bhattacharya RC (2012) Differential expression of salt overly sensitive pathway genes determines salinity stress tolerance in Brassica genotypes. Plant Physiol Biochem 51:90–101. https://doi.org/10.1016/j.plaphy.2011.10.001
Coumou D, Rahmsdorf S (2012) A decade of weather extremes. Nat Clim Chang 2:491–496. https://doi.org/10.1038/nclimate1452
Craita EB, Tom G (2013) Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4:273. https://doi.org/10.3389/fpls.2013.00273
Espinosa-Diez C, Miguel V, Mennerich D, Kietzmann T, Sánchez-Pérez P, Cadenas S, Lamas S (2015) Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol 6:183–197. https://doi.org/10.1016/j.redox.2015.07.008
Frank G, Pressman E, Ophir R, Althan L, Shaked R, Freedman M, Shen S, Firon N (2009) Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J Exp Bot 60(13):3891–3908. https://doi.org/10.1093/jxb/erp234
Fu Y, Yang L, Gao H, Wenji X, Li Q, Li H, Gao J (2020) Comparative transcriptome analysis reveals heat stress-responsive genes and their signalling pathways in lilies (Lilium longiflorum vs. Lilium distichum). PLoS ONE 15(10):e0239605. https://doi.org/10.1371/journal.pone.0239605
Gall HL et al (2015) Cell wall metabolism in response to abiotic stress. Plants (basel) 4(1):112–166. https://doi.org/10.3390/plants4010112
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36(10):3420–3435. https://doi.org/10.1093/nar/gkn176
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, de Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Guo M, Liu J-H, Ma X, Luo D-X, Gong Z-H, Lu M-H (2016) The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front Plant Sci 7:114. https://doi.org/10.3389/fpls.2016.00114
Hong SW, Vierling E (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27(1):25–35. https://doi.org/10.1046/j.1365-313x.2001.01066.x
Hwang JE, Kim YJ, Shin MH, Hyun HJ, Bohnert HJ, Park HC (2018) A comprehensive analysis of the Korean fir (Abies koreana) genes expressed under heat stress using transcriptome analysis. Sci Rep 8(1):1–11. https://doi.org/10.1038/s41598-018-28552-1
Jia XL, Wang GL, Xiong F, Yu XR, Xu ZS, Wang F, Xiong AS (2015) De novo assembly, transcriptome characterization, lignin accumulation and anatomic characteristics: novel insights into lignin biosynthesis during celery leaf development. Sci Rep 5:8259. https://doi.org/10.1038/srep08259
Kanehisa M, Goto S (2000) KEGG: kyotoencyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30. https://doi.org/10.1093/nar/28.1.27
Khan A, Ali M, Khattak AM, Gai WX, Zhang HX, Wei AM, Gong ZH (2019) Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int J Mol Sci 20(21):5321. https://doi.org/10.3390/ijms20215321
Kosar F, Akram NA, Sadiq M, Al-Qurainy F, Ashraf M (2019) Trehalose: a key organic osmolyte effectively involved in plant abiotic stress tolerance. J Plant Growth Regul 38:606–618. https://doi.org/10.1007/s00344-018-9876-x
Kumar RR, Goswami S, Sharma SK, Kala YK, Rai GK, Mishra DC, Grover M, Singh GP, Pathak H, Rai A, Chinnusamy V, Rai RD (2015) Harnessing next generation sequencing in climate change: RNA-seq analysis of heat stress-responsive genes in wheat (Triticum aestivum L.). OMICS 19(10):632–647. https://doi.org/10.1089/omi.2015.0097
Le Nguyen K, Grondin A, Courtois B, Gantet P (2019) Next-generation sequencing accelerates crop gene discovery. Trends Plant Sci 24(3):263–274. https://doi.org/10.1016/j.tplants.2018.11.008
Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25:754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. https://doi.org/10.1093/bioinformatics/btl158
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Li X, Sun H, Pei J, Dong Y, Wang F, Chen H, Sun Y, Wang N, Li H, Li Y (2012) De novo sequencing and comparative analysis of the blueberry transcriptome to discover putative genes related to antioxidants. Gene 511(1):54–61. https://doi.org/10.1016/j.gene.2012.09.021
Li Y, Xu C, Lin X, Cui B, Wu R, Pang X (2014) De novo assembly and characterization of the fruit transcriptome of chinese jujube (Ziziphus jujuba Mill) using 454 pyrosequencing and the development of novel tri-nucleotide SSR markers. PLoS ONE 9(9):e106438. https://doi.org/10.1371/journal.pone.0106438
Liu H, Able AJ, Able JA (2020) Integrated analysis of small rna, transcriptome, and degradome sequencing reveals the water-deficit and heat stress response network in durum wheat. Int J Mol Sci 21(17):6017. https://doi.org/10.3390/ijms21176017
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Ma QH, Wang GX, Liang LS (2011) Development and characterization of SSR markers in Chinese jujube (Ziziphus jujuba Mill) and its related species. Sci Horticult 129(4):597–602. https://doi.org/10.1016/j.scienta.2011.04.032
Malik VS (2006) RNA sequencing as a tool for understanding biological complexity of abiotic stress in plants. J Plant Biochem Biotechnol 25:1–2. https://doi.org/10.1007/s13562-015-0326-6
Mishra RC, Grover A (2016) ClpB/Hsp100 proteins and heat stress tolerance in plants. Crit Rev Biotechnol 36:862–874. https://doi.org/10.3109/07388551.2015.1051942
Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A (2009) Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant PhysiolBiochem 47(9):785–795. https://doi.org/10.1016/j.plaphy.2009.05.003
Nieto-Sotelo J, Martinez LM, Ponce G, Cassab GI, Alagon A, Meeley RB, Ribaut JM, Yang R (2002) Maize HSP101 plays important roles in both induced and basal thermotolerance and primary root growth. Plant Cell 14(7):1621–1633. https://doi.org/10.1105/tpc.010487
Obaid AY, Sabir JSM, Atef A, Liu X, Edris S, El-Domyati FM, Mutwakil MZ, Gadalla NO, Hajrah NH, Al-Kordy MA, Hall N, Bahieldin A, Jansen RK (2016) Analysis of transcriptional response to heat stress in Rhazyastricta. BMC Plant Biol 16:252. https://doi.org/10.1186/s12870-016-0938-6
Ortiz R, Braun HJ, Crossa J, Crouch JH, Davenport G, Dixon J et al (2008) Wheat genetic resources enhancement by the international maize and wheat improvement center (CIMMYT). Genet Resour Crop Evol 55:1095–1140. https://doi.org/10.1007/s10722-008-9372-4
Padaria JC, Yadav R, Tarafdar A, Lone SA, Kumar K, Sivalingam PN (2016) Molecular cloning and characterization of drought stress responsive abscisic acid-stress-ripening (Asr1) gene from wild jujube, Ziziphus nummularia (Burm. f) Wight & Arn. Mol Biol Rep 43(8):849–859. https://doi.org/10.1007/s11033-016-4013-z
Pandey A, Singh R, Radhamani J, Bhandari DC (2010) Exploring the potential of Ziziphus nummularia (Burm. F.) Wight et Arn. from drier regions of India. Genetic Resour Crop Evolut 57(6):929–936. https://doi.org/10.1007/s10722-010-9566-4
Panzade KP, Vishwakarma H, Padaria JC (2020) Heat stress inducible cytoplasmic isoform of ClpB 1 from Z. nummularia exhibits enhanced thermotolerance in transgenic tobacco. Mol Biol Rep 47(5):3821–3831. https://doi.org/10.1007/s11033-020-05472-w1-11
Planton S, Driouech F, Rhaz KE, Lionello P (2016) The climate of the mediterranean regions in the future climate projections. In : the mediterranean region under climate change: a scientific update [enligne]. Marseille: IRD Éditions, 2016 (généré le 12 septembre 2022). ISBN: 9782709922203. https://doi.org/10.4000/books.irdeditions.23085
Pollock K, Yu G, Moller-Trane R, Koran M, Dosa PI, McKenna DH, Hubel A (2016) Combinations of osmolytes, including monosaccharides, disaccharides, and sugar alcohols act in concert during cryopreservation to improve mesenchymal stromal cell survival. Tissue Eng Part C Methods 22(11):999–1008. https://doi.org/10.1089/ten.TEC.2016.0284
Pradhan SK, Pandit E, Nayak DK, Behera L, Mohapatra T (2019) Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol 19(1):352. https://doi.org/10.1186/s12870-019-1922-8
Qu AL, Ding YF, Jiang Q, Zhu C (2013) Molecular mechanisms of the plant heat stress response. Biochem Biophys Res Commun 432(2):203–207. https://doi.org/10.1016/j.bbrc.2013.01.104
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Sagor GHM, Berberich T, Takahashi Y, Niitsu M, Kusano T (2013) The polyamine spermine protects Arabidopsis from heat stress-induced damage by increasing expression of heat shock-related genes. Transgenic Res 22(3):595–605. https://doi.org/10.1007/s11248-012-9666-3
Schirmer EC, Glover JR, Singer MA, Lindquist S (1996) HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci 21(8):289–296
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31(3):279–292. https://doi.org/10.1046/j.1365-313x.2002.01359.x
Shi H, Chan Z (2014) Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J Integr Plant Biol 56(2):1672–9072. https://doi.org/10.1111/jipb.12128
Shi CY, Yang H, Wei CL et al (2011) Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 12(1):131. https://doi.org/10.1186/1471-2164-12-131
Singh AK, Devanshi Sharma P, Singh R, Singh B, Koundal K, Singh NK (2007) Assessment of genetic diversity in Ziziphus mauritianausing inter-simple sequence repeat markers. J Plant Biochem Biotechnol 16:35–40. https://doi.org/10.1007/BF03321926
Tenhaken R (2014) Cell wall remodeling under abiotic stress. Front Plant Sci 5:771. https://doi.org/10.3389/fpls.2014.00771
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422. https://doi.org/10.1007/s00122-002-1031-0
Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61:199–223. https://doi.org/10.1016/j.envexpbot.2007.05.011
Wang Z, Fang B, Chen J, Zhang X, Luo Z, Huang L, Chen X (2010) De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweetpotato (Ipomoea batatas). BMC Genomics 11:726. https://doi.org/10.1186/1471-2164-11-726
Wang S, Wei XL, Cheng LJ, Tong ZK (2014) Identification of a C2H2-type zinc finger gene family from Eucalyptus grandis and its response to various abiotic stresses. Biol Plant 58:385–390. https://doi.org/10.1007/s10535-014-0399-4
Wang Y, Dong C, Xue Z, Jin Q, Xu Y (2016) De novo transcriptome sequencing and discovery of genes related to copper tolerance in Paeonia ostii. Gene 576(1):126–135. https://doi.org/10.1016/j.gene.2015.09.077
Xia Z, Xu H, Zhai J, Li D, Luo H, He C, Huang X (2011) RNA-Seq analysis and de novo transcriptome assembly of Hevea brasiliensis. Plant Mol Biol 77(3):299–308. https://doi.org/10.1007/s11103-011-9811-z
Xu P, Liu Z, Fan X, Gao J, Zhang X, Zhang X, Shen X (2013) De novo transcriptome sequencing and comparative analysis of differentially expressed genes in Gossypium aridum under salt stress. Gene 525(1):26–34. https://doi.org/10.1016/j.gene.2013.04.066
Xu H, Chen L, Song B, Fan X, Yuan X, Chen J (2016) De novo transcriptome sequencing of pakchoi (Brassica rapa L. chinensis) reveals the key genes related to the response of heat stress. Acta Physiol Plant 38:252. https://doi.org/10.1007/s11738-016-2269-5
Yadav R, Lone SA, Gaikwad K et al (2018a) Transcriptome sequence analysis and mining of SSRs in Jhar Ber (Ziziphus nummularia (Burm.f.) Wight & Arn) under drought stress. Sci Rep 8:2406. https://doi.org/10.1038/s41598-018-20548-1
Yadav R, Verma OP, Padaria JC (2018b) Transcript profiling and gene expression analysis under drought stress in Ziziphus nummularia (Burm.f.) Wright & Arn. Mol Biol Rep 45:163–174. https://doi.org/10.1007/s11033-018-4149-0
Yan J, Yu L, Xuan J, Lu Y, Lu S, Zhu W (2016) De novo transcriptome sequencing and gene expression profiling of spinach (Spinacia oleracea L.) leaves under heat stress. Sci Rep 9(6):19473. https://doi.org/10.1038/srep19473
Zhang C, Yin X, Li X, Huang J, Wang C, Lian C (2013) Development and characterization of microsatellite markers for sour jujube (Ziziphus jujuba var. spinosa) Indian. J Genet Plant Breed 73(03):338–341. https://doi.org/10.5958/j.0975-6906.73.3.052
Acknowledgements
We sincerely acknowledge Director, ICAR–National Institute for Plant Biotechnology, New Delhi 110012, Delhi, India for providing support and facilities to carry out this research work. We also gratefully acknowledge the Director, ICAR-Indian Agricultural Research Institute (IARI) for providing National Phytotron Facility (NPF), Delhi 110012, India, for providing the plant growth chamber to conduct the research under controlled conditions. Fellowship received from Department of Biotechnology (DBT), Government of India by the first author is duly acknowledged.
Funding
No external fund grant was received to carry out this work.
Author information
Authors and Affiliations
Contributions
JCP conceptualized and designed the study. KPP conducted the experiments, analysed and interpreted the results and wrote the manuscript under the supervision of JCP. KUT, DVP analyzed the data and edited the manuscript. KUT submitted the datasets to Sequence Read Archive (SRA) repository. RSJ involved manuscript editing. KG, MD, RR, MPS, OPA joined discussions regarding the experiments and data interpretations. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors of this manuscript have no competing interests. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Panzade, K.P., Tribhuvan, K.U., Pawar, D.V. et al. Discovering the regulators of heat stress tolerance in Ziziphus nummularia (Burm.f) wight and walk.-arn. Physiol Mol Biol Plants 30, 497–511 (2024). https://doi.org/10.1007/s12298-024-01431-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-024-01431-y