Abstract
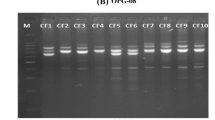
Picrorhiza kurroa is a medicinally important, high altitude perennial herb, endemic to the Himalayas. It possesses strong hepato-protective bioactivity that is contributed by two iridoid picroside compounds viz Picroside-I (P-I) and Picroside-II (P-II). Commercially, many P. kurroa based hepato-stimulatory Ayurvedic drug brands that use different proportions of P-I and P-II are available in the market. To identify genetically heterozygous and high yielding genotypes for multiplication, sustained use and conservation, it is essential to assess genetic and phytochemical diversity and understand the population structure of P. kurroa. In the present study, isolation and HPLC based quantification of picrosides P-I and P-II and molecular DNA fingerprinting using RAPD, AFLP and ISSR markers have been undertaken in 124 and 91 genotypes, respectively. The analyzed samples were collected from 10 natural P. kurroa Himalayan populations spread across four states (Jammu & Kashmir, Sikkim, Uttarakhand and Himachal Pradesh) of India. Genotypes used in this study covered around 1000 km geographical area of the total Indian Himalayan habitat range of P. kurroa. Significant quantitative variation ranging from 0.01 per cent to 4.15% for P-I, and from 0.01% to 3.18% in P-II picroside was observed in the analyzed samples. Three molecular DNA markers, RAPD (22 primers), ISSR (15 primers) and AFLP (07 primer combinations) also revealed a high level of genetic variation. The percentage polymorphism and effective number of alleles for RAPD, ISSR and AFLP analysis varied from 83.5%, 80.6% and 72.1%; 1.5722, 1.5787 and 1.5665, respectively. Further, the rate of gene flow (Nm) between populations was moderate for RAPD (0.8434), and AFLP (0.9882) and comparatively higher for ISSR (1.6093). Fst values were observed to be 0.56, 0.33, and 0.51 for RAPD, ISSR and AFLP markers, respectively. These values suggest that most of the observed genetic variation resided within populations. Neighbour joining (NJ), principal coordinate analysis (PCoA) and Bayesian based STRUCTURE grouped all the analyzed accessions into largely region-wise clusters and showed some inter-mixing between the populations, indicating the existence of distinct gene pools with limited gene flow/exchange. The present study has revealed a high level of genetic diversity in the analyzed populations. The analysis has resulted in identification of genetically diverse and high picrosides containing P. kurroa genotypes from Sainj, Dayara, Tungnath, Furkia, Parsuthach, Arampatri, Manvarsar, Kedarnath, Thangu and Temza in the Indian Himalayan region. The inferences generated in this study can be used to devise future resource management and conservation strategies in P. kurroa.












Similar content being viewed by others
Availability of data materials
Available on request.
References
Akbar S (2020) Picrorhiza kurroa Royle ex Benth. (Plantaginaceae). In: Handbook of 200 medicinal plants: a comprehensive review of their traditional medical uses and scientific justifications. Springer, Cham. https://doi.org/10.1007/978-3-030-16807-0_145
Barik SK, Rao BR, Haridasan K, Adhikari D, Singh PP, Tiwary R (2018) Classifying threatened species of India using IUCN criteria. Curr Sci 114:588–595
Barrett SCH, Kohn JK (1991) Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KH (eds) Genetics and conservation of rare plants. Center for plant conservation. Oxford University Press, Oxford, pp 3–30
Belaj A, Satovic Z, Cipriani G, Baldoni L, Testolin R, Rallo L, Trujillo I (2003) Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theor Appl Genet 107:736–744
Bhat WW, Dhar N, Razdan S, Rana S, Mehra R, Nargotra A et al (2013) Molecular characterization of UGT94F2 and UGT86C4, two Glycosyltransferases from Picrorhiza kurrooa: comparative structural insight and evaluation of substrate recognition. PLoS ONE 8(9):e73804. https://doi.org/10.1371/journal.pone.0073804
Bhattacharjee S, Bhattacharya S, Jana S, Baghel DS (2013) A review on medicinally important species of Picrorhiza. IJPRBS 2(4):1–16
Carvalho YGS, Vitorino LC, de Souza UJB, Bessa LA (2019) Recent trends in research on the genetic diversity of plants: implications for conservation. Diversity. https://doi.org/10.3390/d11040062
Chesnokov Y, Artem’eva A (2015) Evaluation of the measure of polymorphism. Agric Biol 50(5):571–578
Costa R, Pereira G, Garrido I, Tavares-de-Sousa MM, Espinosa F (2016) Comparison of RAPD, ISSR, and AFLP molecular markers to reveal and classify Orchardgrass (Dactylis glomerata L.) germplasm variations. PLoS ONE 11(4):e0152972. https://doi.org/10.1371/journal.pone.0152972
De Masi L, Siviero P, Esposito C, Castaldo D, Siano F, Laratta B (2006) Assessment of agronomic, chemical and genetic variability in common basil (Ocimum basilicum L.). Eur Food Res Technol 223(2):273. https://doi.org/10.1007/s00217-005-0201-0
Debnath P, Rathore S, Walia S, Kumar M, Devi R, Kumar R (2020) Picrorhiza kurroa: a promising traditional therapeutic herb from higher altitude of western Himalayas. J Herb Med 23:100358. https://doi.org/10.1016/j.hermed.2020.100358
Degani C, Rowland LJ, Saunders AJA, Hokanson SC, Ogden EL, Golan-Goldhirsh A, Galletta GJ (2001) A comparison of genetic relationship measures in strawberry (Fragaria x ananassa Duch.) based on AFLPs, RAPDs, and pedigree data. Euphytica 117:1–12
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.1: An integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Hadian J, Ebrahimi SN, Mirjalili MH, Azizi A, Ranjbar H, Friedt W (2011) Chemical and genetic diversity of Zataria multiflora Boiss. accessions growing wild in Iran. Chem Biodiver 8:176–188
Hamrick JL, Godt MJW (1990) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding and genetic resources. Sinauer, Sunderland, pp 43–63
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond B Biol Sci 351:1291–1298
Han T, Hu Y, Zhou SY et al (2008) Correlation between the genetic diversity and variation of total phenolic acids contents in Fructus xanthii from different populations in China. Biomed Chromatogr 22:478–486
Katoch M, Hussain MA, Ahuja A (2013) Comparison of SSR and cytochrome P-450 markers for estimating genetic diversity in Picrorhiza kurrooa L. Plant Syst Evol 299:1637–1643
Katoch M, Fazli IS, Suri KA et al (2011) Effect of altitude on picroside content in core collections of Picrorhiza kurrooa from the north western Himalayas. J Nat Med 65:578–582. https://doi.org/10.1007/s11418-010-0491-9
Krishnamurthy A (1969) The Wealth of India, vol VIII. Publication and Information Directorate, CSIR, New Delhi, p 49
Kumar A, Rajpal VR, Raina R, Choudhary M, Raina SN (2014) Nuclear DNA assay of the wild endangered medicinal and aromatic Indian Himalayan Valeriana jatamansi germplasm with multiple DNA markers: implications for genetic enhancement, domestication and ex situ conservation. Plant Syst Evol 300(9):2061–2071
Kumar J, Verma V, Goyal A, Shahi AK, Saproo R, Sangwan RS, Qazi GN (2009) Genetic diversity analysis in Cymbopogon species using DNA markers. Plant Omics 2(1):20
Kumar N, Sharma SK (2014) A new aromatic ester from Picrorhiza kurroa Royle ex Benth. J Pharmacogn Phytochem 3(1):96–98
Kumar V (2019) OMICS‐Based approaches for elucidation of picrosides biosynthesis in Picrorhiza Kurroa. https://doi.org/10.1002/9781119509967.ch8
Kumar V, Bansal A, Chauhan RS (2017) Modular design of picroside-II biosynthesis deciphered through NGS transcriptomes and metabolic intermediates analysis in naturally variant chemotypes of a medicinal herb, Picrorhiza kurroa. Front Plant Sci 8:564. https://doi.org/10.3389/fpls.2017.00564
Kumar V, Sharma N, Sood H et al (2016) Exogenous feeding of immediate precursors reveals synergistic effect on picroside-I biosynthesis in shoot cultures of Picrorhiza kurroa Royle ex Benth. Sci Rep 6:29750
Lannér-Herrera C, Gustafsson M, Fält AS, Bryngelsson T (1996) Diversity in natural populations of wild Brassica oleracea as estimated by Isozyme and RAPD analysis. Genet Resour Crop Evol 43:13–23
Lone SA, Qazi PH, Gupta S (2018) Genetic diversity of Epimedium elatum (Morren & Decne) revealed by RAPD characterization. Curr Bot 9:41–46
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Mahajan R, Kapoor N, Singh I (2016) Somatic embryogenesis and callus proliferation in Picrorhiza kurroa Royle ex. Benth J Exp Biol Agric Sci 4(2):201–209
Mandal S, Mukhopadhyay S (2004) New iridoid glucoside from Picrorhiza kurroa Royle ex Benth. Indian J Chem Sect B Org Chem Incl Med Chem 43(5):1023–1025
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Can Res 27:209–220
Masood M, Arshad M, Qureshi R, Sabir S, Amjad MS, Qureshi H, Tahir Z (2015) Picrorhiza kurroa: An ethnopharmacologically important plant species of Himalayan region. Pure Appl Biol 4(3):407–417
Mehra TS, Raina R, Rana RC, Chandra P, Pandey PK (2017) Elite chemo-types of a critically endangered medicinal plant Picrorhiza kurroa Royle ex Benth from Indian Western Himalaya. J Pharmacogn Phytochem 6(5):1679–1682
Naik PK, Alam MA, Singh H, Goyal V, Parida S, Kalia S, Mohapatra T (2010) Assessment of genetic diversity through RAPD, ISSR and AFLP markers in Podophyllum hexandrum: a medicinal herb from the Northwestern Himalayan region. Physiol Mol Biol Plants 16(2):135–148. https://doi.org/10.1007/s12298-010-0015-9
Nayar MP, Sastry ARK (1990) Red data plants of India. CSIR Publication, New Delhi, p 271
Nazarzadeh Z, Onsori H, Akrami S (2020) Genetic diversity of bread wheat (Triticum aestivum L.) genotypes using RAPD and ISSR molecular markers. J Genet Resour 6(1):69–76
Pejic I, Ajmone-Marsan P, Morgante M, Kozumplik V, Castiglioni P, Taramino G, Motto M (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs and AFLPs. Theor Appl Genet 97:1248–1255
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Prentice HC, Malm JU, Mateu-Andres I, Segarra-Moragues JG (2003) Allozyme and chloroplast DNA variation in island and mainland populations of the rare Spanish endemic, Silene hifacensis (Caryophyllaceae). Conserv Genet 4:543–555
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rohlf FJ (1998) NTSYS-pc: numerical taxonomy and multivariate analysis system version 2.02K. Applied Biostatistics, New York
Russell JR, Fuller JB, MaCaulay M, Hatz BG, Jhoor A, Powel W, Waugh R (1997) Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs. Theor Appl Genet 95:1161–1168
Sales E, Sergio GN, Mus M, Segura J (2001) Population genetic study in the Balearic endemic plant species Digitalis minor (Scrophulariaceae) using RAPD markers. Am J Bot 88(10):1750–1759
Sarwat M, Das S, Srivastava PS (2008) Analysis of genetic diversity through AFLP, SAMPL, ISSR and RAPD markers in Tribulus terrestris, a medicinal herb. Plant Cell Rep 27:519–528
Shitiz K, Pandit S, Chanumolu S, Sood H, Singh H, Singh J, Chauhan R (2017) Mining simple sequence repeats in Picrorhiza kurroa transcriptomes for assessing genetic diversity among accessions varying for picrosides contents. Plant Genet Resour 15(1):79–88
Shitiz K, Sharma N, Pal T, Sood H, Chauhan RS (2015) NGS Transcriptomes and enzyme inhibitors unravel complexity of picrosides biosynthesis in Picrorhiza kurroa Royle ex. Benth PLoSONE 10(12):e0144546
Singh P, Nag A, Parmar R, Ghosh S, Bhau BS, Sharma RK (2015) Genetic diversity and population structure of endangered Aquilaria malaccensis revealed potential for future conservation. J Genet 94:697–704
Singh P, Sharma RK (2020) Development of informative genic SSR markers for genetic diversity analysis of Picrorhiza kurroa. J Plant Biochem Biotechnol 29:144–148
Smith JF, Pham TV (1996) Genetic diversity of the narrow endemic Allium aaseae (Alliaceae). Am J Bot 83:717–726
Song BH, Varshney VK, Mittal N, Ginwal HS (2015) High levels of diversity in the phytochemistry, ploidy and genetics of the medicinal plant Acorus calamus L. Med Aromat Plants S 1:002. https://doi.org/10.4172/2167-0412.S1-002
Soni D, Grover A (2019) “Picrosides” from Picrorhiza kurroa as potential anti-carcinogenic agents. Biomed Pharmacother 109:1680–1687. https://doi.org/10.1016/j.biopha.2018.11.048
Sultan P, Jan A, Pervaiz Q (2016) Phytochemical studies for quantitative estimation of iridoid glycosides in Picrorhiza kurroa Royle. Bot Stud 57:7. https://doi.org/10.1186/s40529-016-0121-2
Sultan P, Shawl AS, Ramtekeb PW, Koura A, Qazia PH (2008) Assessment of diversity in Podophyllum hexandrum by genetic and phytochemical markers. Sci Hortic 115:398–408
Thakur RK, Rajpal VR, Raina SN, Kumar P, Sonkar A, Joshi L (2020) UPLC-DAD assisted phytochemical quantitation reveals a sex, ploidy and ecogeography specificity in the expression levels of selected secondary metabolites in medicinal Tinospora cordifolia: implications for elites’ identification program. Curr Top Med Chem 20:698. https://doi.org/10.2174/1568026620666200124105027
Thani PR, Sharma YP, Kandel P (2018) Phytochemical studies on Indian market samples of drug “Kutki” (Picrorhiza kurroa Royle ex Benth). Re J Agr Forest Sci 6(2):1–5
Thapliyal S, Mahadevan N, Nanjan MJ (2012) Analysis of Picroside I and Kutkoside in Picrorhiza kurroa and its Formulation by HPTLC. Int J Res Pharm Biomed Sci 3(1):25–30
Thein SI, Wallace RR (1986) The use of synthetic oligonucleotides as specific hybridization probes in the diagnosis of genetic disorders. In: Davis KE (ed) Human genetic disease: a practical approach. IRL, Oxford, pp 33–50
Thiyagarajan M, Venkatachalam P (2015) Assessment of genetic and biochemical diversity of Stevia rebaudiana Bertoni by DNA fingerprinting and HPLC analysis. Ann Phytomed 4(1):79–85
Tiwari SS, Pandey MM, Srivastava S, Rawat A (2012) KTLC densitometric quantification of picrosides (picroside-I and picroside-II) in Picrorhiza kurroa and its substitute Picrorhiza scrophulariiflora and their antioxidant studies. Biomed Chromatogr 26:61–68
Torres E, Iriondo JM, Pérez C (2003) Genetic structure of an endangered plant, Antirrhinum microphyllum (Scrophulariaceae): Allozyme and RAPD analysis. Am J Bot 90:85–92
Trindade H, Costa MM, Sofia BLA, Pedro LG, Figueiredo AC, Barroso JG (2008) Genetic diversity and chemical polymorphism of Thymus caespititius from Pico, Sao Jorge and Terceira islands (Azores). Biochem Syst Ecol 36(10):790–797
Vaidya AB, Antarkar DS, Doshi JC, Bhatt AD, Ramesh V, Vora PV, Perissond D, Baxi AJ, Kale PMJ (1996) Picrorhiza kurroa (Kutki) Royle ex Benth as a hepatoprotective agent experimental and clinical studies. J Postgrad Med 42:105–108
Yeh FC, Boyle T, Yeh Z, Xiyan JM (1999) POPGENE Version 1.31: Microsoft Window based freeware for population genetic analysis. University of Alberta and center for International Forestry Research, Edmonton.
Acknowledgements
This work was supported by grant for scientific research from the Department of Biotechnology, Government of India. Avinash Kumar acknowledges SRF from CSIR during his Ph.D. The authors are thankful to Prof. (Retd.) Ravinder Raina, Department of Forest Products, Dr. Y. S. Parmar University of Horticulture and Forestry, Solan, Himachal Pradesh, India, for easing collection of germplasm from Himachal. The authors extend their sincere gratitude to anonymous reviewers for their constructive comments that helped in the improvement of the manuscript.
Funding
This work was supported by grant for scientific research from the Department of Biotechnology, Government of India.
Author information
Authors and Affiliations
Contributions
Contributors Role Taxonomy (CRediT): Avinash Kumar: Sample Collection, Methodology, Investigation (molecular profiling), Validation, Formal analysis, Writing—Original Draft preparation. Ambika: Methodology, Investigation (Chemical profiling), Formal analysis, Validation, (chemoprofiling). Amita Kumari: Analysis. Rachhaya.Mallikarjun Devarumath: Sample Collection. Rakesh Thakur: Analysis. Manju Chaudhary: Analysis. Pradeep Pratap Singh: Analysis (Chemical profiling). Shiv Murat Singh Chauhan: Conceptualization, Supervision (Chemical profiling work). SN Raina: Conceptualization, Resources, Overall Supervision, Project administration, Funding acquisition. Vijay Rani Rajpal: Supervision, Visualization, Validation, Writing and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
All authors have given their consent for publication in the journal “Physiology and Molecular Biology of Plants”.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, A., Rajpal, V.R., Ambika et al. Isolation and HPLC assisted quantification of two iridoid glycoside compounds and molecular DNA fingerprinting in critically endangered medicinal Picrorhiza kurroa Royle ex Benth: implications for conservation. Physiol Mol Biol Plants 27, 727–746 (2021). https://doi.org/10.1007/s12298-021-00972-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-00972-w