Abstract
Infections caused by the genus Fusarium have emerged over the past decades and range from onychomycosis and keratitis in healthy individuals to deep and disseminated infections with high mortality rates in immune-compromised patients. As antifungal susceptibility can differ between the different Fusarium species, identification at species level is recommended. Several clinical observations as hyaline hyphae in tissue, necrotic lesions in the skin and positive blood tests with fungal growth or presence of fungal cell wall components may be the first hints for fusariosis. Many laboratories rely on morphological identification, but especially multi-locus sequencing proves better to discriminate among members of the species complexes involved in human infection. DNA-based diagnostic tools have best discriminatory power when based on translation elongation factor 1-α or the RNA polymerase II second largest subunit. However, assays based on the detection of other fusarial cell compounds such as peptides and cell wall components may also be used for identification. The purpose of this review is to provide an overview and a comparison of the different tools currently available for the diagnosis of fusariosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Taking the genus Fusarium in its currently broadest definition [1•], it contains many species with either saprophytic or plant pathogenic lifestyles. However, the genus also contains species capable of forming a plethora of mycotoxins and, more importantly, harbours species capable of causing opportunistic infections in human and animals [2, 3•]. In recent years, the numbers of infections reported to be caused by Fusarium species have increased [4, 5].
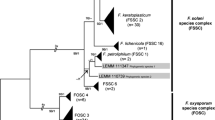
Many species within the genus Fusarium that were recognized based on morphological characters proved to be species complexes, with little to no morphological differences, rather than single species. For their recognition, often multi-locus sequence typing (MLST) is required [6–9]. Most of the identified opportunistic Fusarium pathogens belong to the Fusarium solani species complex (FSSC), the Fusarium oxysporum species complex (FOSC) and Fusarium fujikuroi species complex (FFSC). Less frequently encountered are members of the Fusarium incarnatum-equiseti (FIESC), Fusarium dimerum (FDSC) and Fusarium chlamydosporum species complexes (FCSC) or species such as Fusarium sporotrichioides [8, 10, 11]. Some of these species cause infections worldwide, but other species appear to be endemic in certain areas [12, 13].
Fusarium species cause a broad spectrum of opportunistic infections in humans. In otherwise healthy individuals, the most prevalent fusarioses are onychomycosis and skin infections—that due to their lack of urgency often go undiagnosed—and keratitis that especially in warmer, drier climates can reach high incidences. In immunocompromised patients, deep invasion at the primary site of infection can occur, while especially in patients with haematological disorders, disseminated infections occur [3•, 14]. In the case of disseminated fusariosis, the manifestation of multiple necrotic lesions spread over the body and positive blood cultures are highly suggestive of fusariosis.
Fusarium strains have high levels of intrinsic antifungal resistance: Recently, diagnostic guidelines recommend amphotericin B (AMB) and voriconazole (VOR) as the preferred drugs of choice for treatment of deep and disseminated infections and recommend against the use of echinocandins [15•]. Although AMB and VOR are sufficient for the treatment of the majority of Fusarium infections, some Fusarium species are not susceptible to AMB and VOR or posaconazole (POS) [16, 17]. In many countries, fungal keratitis is treated with natamycin, that is effective against most but not all, Fusarium species, e.g. [18]. Detailed studies on antifungal susceptibility profiles emphasize the need for species-level identification for best-adapted treatment strategies [5].
This review gives an overview of the currently available techniques for the diagnosis of fusarioses and for the identification of Fusarium as opposed to other fungal species and within the species to species complex or preferably species level. Here, we also discuss whether each diagnostic technique is suitable for direct detection of Fusarium in patient specimens or whether it is restricted to identification of a cultured isolate.
Clinical Observations Indicative for Fusariosis
Some clinical observations may give a first hint to Fusarium being the etiological agent causing an infection:
Direct Microscopy in Keratitis
In vivo confocal microscopy of the infected eye is a non-invasive and rapid technique to determine whether there is a fungal or other microbial infection and has become part of the routine especially in countries where there is a high incidence of fungal keratitis [19–21]. Sensitivity and specificity of fungal and acanthamoebal diagnosis by confocal microscopy are estimated at 88.3 and 91.1 %, respectively [21]. Even the modern smartphone camera and pocket magnifier can now be used to detect fungal hyphae in corneal scrapings [22]. However, fungal structures that enable genus or even species-level identification are seldom formed in clinical specimens [20].
Blood Tests in Disseminated Infections
Members of the genus Fusarium are among the few filamentous fungal species yielding positive blood cultures, while it is known for decades that such disseminated fusariosis is often accompanied by multiple necrotic skin lesions [23, 24]. Less well known is the fact that the serum galactomannan tests developed for Aspergillus infections can also cross-react with Fusarium infections, since both genera have a similar cell wall structure and hence may be an indication for disseminated fusariosis [16, 25]. The Platelia Candida antigen detection enzyme immunoassay directed against mannan has been reported to give cross-reactions with Fusarium verticillioides, but not with F. solani or F. oxysporum [26]. Also the presence of beta-glucan is not specific for particular fungal genera, and Fusarium spp. do produce this cell wall component [27].
Radiology
Radiological findings may be helpful for the identification of infective fungal diseases, but they are often not specific for a given species/etiological agent, as many times the findings include alveolar and interstitial infiltrates, nodules and cavities that are typical for a wide range of diseases. Studying pulmonary fusariosis, Marom et al. [28] showed that nodules and lung masses are seen in 82 % of the patients on CT scans but only in 45 % on chest radiography. Non-specific findings were observed in 30 % of the patients, while findings at presentation were observed in 25 %. Halo and reverse halo signs are absent in Fusarium infections [28, 29]. Early chest CT imaging in immunocompromised patients suspected of having invasive fungal pneumonia can help identify disease early, leading to improved outcome [30].
Histology/immunohistochemistry
Confirmatory diagnosis of fusariosis by histopathology is strongly recommended by the European Confederation of Medical Mycology and European Society of Clinical Microbiology and Infectious Diseases (ECMM/ESCMID) guidelines [15•]. In tissue, the hyphae of Fusarium are similar to those for instance Aspergillus species, with hyaline and septate filaments that typically dichotomize in acute and right angles [14]. Adventitious yeast-like sporulation may be present in tissue, and the finding of hyphae and yeast-like structures together is highly suggestive of fusariosis in the high-risk population [14]. Some Fusarium species may form chlamydospores in the hyphae in tissue [31]. However, overall, directly observed morphological characteristics in histopathological examination are non-specific.
Alternate techniques have been used to determine the specific agent: Immunohistochemistry uses antibodies directed at fungal cell antigens—mainly proteins—to identify organisms visualized in tissue. Advantages of immunohistochemistry include relatively rapid results, preservation of tissue morphology and the ability to use formalin-fixed paraffin-embedded (FFPE) tissue. However, few antibodies are commercially available for specific moulds [32], but some authors have described the more or less specific detection of Fusarium in human tissue with in-house-made polyclonal fluorescent or peroxidase-linked antibodies, e.g. [33, 34].
In the case of in situ hybridization (ISH), the target for recognition is DNA or RNA, while as probe, a labelled complementary strand is used. Several types of probes have been successfully used for the detection of Fusarium strains: oligonucleotide DNA [35, 36], PNA (peptide nucleic acids; DNA mimics with a peptide backbone) [37, 38] and LNA (a mix of DNA and locked nucleic acid (LNA)-modified nucleotides in which the 2′ oxygen and the 4′ carbon are linked through a methylene unit) [39]. Especially, the PNA and LNA nucleotides hybridize strongly to their complementary RNA and DNA nucleotides, producing hybrids that are thermally stable. Probes can be radio-, fluorescent- or antigen-labelled. Most probes developed for Fusarium species have been directed at the ribosomal RNA and have low levels of specificity.
Once fungal elements have been detected in patient tissue, there is also the option to use PCR-based tools (see below) for their identification. A restriction here is that sufficient pure fungal DNA needs to be isolated from the sample. Several commercial kits are available to isolate DNA from FFPE tissues where deparaffinization can be obtained via xylene or microwave irradiation [40]. Lau et al. [41] provide an example of successful pan-fungal PCR and sequencing both in fresh and FFPE tissue specimens from patients with IFI. Laser capture microdissection (LCM) can allow for the microscopic procurement of specific cell types from FFPE tissue sections for specific further analyses [42].
Morphology-based Identification
Identification of fungal isolates has long been based on morphological characteristics of cultures, and for many laboratories, this is still the standard for identification. However, many Fusarium species look similar in culture and have been shown to represent species complexes instead of single species. Often colonies obtained from patients prove degenerate in their colony morphology or pionnotal [2, 43]. The media and growth conditions used influence growth rate and colony pigmentation, formation of differentiating structures and the dimensions of these structures. The formation of specific structures may take weeks and may require UV or daylight induction to be formed at all.
The Atlas of Clinical Fungi [43] provides a dichotomous key for the morphological identification of clinical fungi till date, including Fusarium. Guarro and Gené in their 1992 paper [44] detailed about the differences between Fusarium, Acremonium and Cylindrocarpon, but Summerbell and Schroers [45] proved some Acremonium and Cylindrocarpon species are actually also members of the genus Fusarium. Azor et al. [46] provide a key with morphological features for clinical Fusarium species that agrees well with molecular identification based on sequences of the beta-tubulin gene. For the identification of Fusarium species in general—not just known clinical strains—Leslie and Summerell’s text [2] provides a convenient tool. Each key uses different preferred media.
DNA-based Identification Techniques
PCR-based
PCR assays are the most widely used method for the amplification of DNA in laboratories. The exponential amplification of target DNA makes it possible to detect even small amounts of fungus, while target choice makes a test more or less specific. Many PCR-based tools have been used for the identification of fungal infections, but only few tools are commercially available, and they depend on in-house validation. Recent guidelines [15•] recommend the use PCR-based diagnostic tools when available for confirmation of infection.
A PCR detection based on the intergenic spacer (IGS) region has been developed for different agricultural important Fusarium species (complexes) that can also distinguish clinical species complexes like Fusarium equiseti and F. sporotrichioides because different-sized fragments are produced [47]. A fluorescent PCR fragment length analysis based on the internally transcribed spacer 2 (ITS2) region is available to distinguish between Aspergillus, Candida and F. oxysporum [48], but the method cannot distinguish between the Fusarium spp. that have (near) identical ITS amplicon lengths. Utilization of a semi-nested PCR (nPCR) increases the sensitivity of this method and makes it usable in blood samples with a low pathogen load [48]. Both Ahmad et al. [49] and Sugawara et al. [50] used nPCR—based on the ribosomal ITS1-5.8S-ITS2 region—to detect Fusarium in bronchoalveolar lavage (BAL), serum and/or blood samples. However, for both authors’ research, the chosen DNA region limits identification to genus level.
In rep-PCR, non-coding repetitive extragenic palindromic (REP) sequences interspersed throughout the fungal genome are amplified using PCR. Initial discovery of REP elements occurred in the genomes of Escherichia coli and Salmonella. REP elements are generally between 33 and 40 bp in length, and the amplified DNA fragments, when separated by electrophoresis, constitute a genomic fingerprint that can be employed for subspecies discrimination and strain delineation of bacteria and fungi. DiversiLab is a commercial automated repetitive sequence-based PCR method that has been tested for a set of 26 Fusarium isolates, only two thirds of which could be identified similarly to sequence-based identification on the large ribosomal subunit (LSU) and elongation factor 1-α [51].
In multiplex PCR, several DNA targets are amplified simultaneously. To distinguish between different genera of human pathogens on one side and opportunists like Candida, Cryptococcus and Fusarium on the other, a multiplex tandem PCR on cultures was developed based on ITS, beta-tubulin and elongation factor 1-α regions [52]. A multiplex PCR based on one gene, galactose oxidase B (gaoB), is available for the differentiation to the species level of several Fusarium maize pathogens that are in fact also human opportunists including F. verticillioides and Fusarium subglutinans [53].
During real-time or quantitative PCR (qPCR), the target DNA is amplified and simultaneously detected, shortening time to diagnosis. The two most commonly used methods act via non-specific dyes intercalating in the double-stranded DNA or via fluorescently labelled probes. The number of cycles required to detect a signal is an indication of the initial amount of target sequence in the sample. Bernal-Martínez et al. [54] used the ribosomal DNA region for development of a duplex qPCR capable of detecting F. solani and non-F. solani species. Testing in a murine model showed high sensitivity and low detection limits, though better for F. solani than for non-solani species tested. Muraosa et al. [55] used a cycling probe technology-based real-time PCR with a chimaera probe, composed of RNA and DNA targeting the 28S ribosomal DNA (rDNA), for the genus Fusarium as well as for FSSC that had a high sensitivity and also worked in serum samples, but not in whole blood samples.
Quantitative multiplex PCRs with labelled primers based on 5.8S and 28S rDNA genes have been described to distinguish between genera of several pulmonary fungal pathogens including Fusarium, Aspergillus and Mucorales. The essay worked both in isolates and different patient samples [56]. A second multiplex real-time PCR method uses hybridization probes for the detection and the quantification of plant and human pathogenic Fusarium proliferatum, F. subglutinans, Fusarium temperatum and F. verticillioides [57].
Furthermore, there are PCR-based tools that are not described for clinical diagnostics such as, e.g. multiplex ligation-dependent probe amplification (MLPA). For studies on the epidemiology, biodiversity and genotyping, amplified fragment length polymorphism (AFLP) analyses [58–61], enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) [61, 62], PCR restriction fragment length polymorphism (PCR-RFLP) [62] and random amplification of polymorphic DNA [60] have been used. However, for diagnostics, these methods would only be applicable with a large enough and validated database.
Hybridization-based Techniques
In reverse line blot (RLB) hybridization, a DNA sample is first subjected to (multiplex) PCR and then screened against a set of specific probes linked to a membrane. Wang and colleagues [63] developed a set of primers based on the IGS region of clinically important Fusarium strains. Their probe set contained one genus-specific, 13 species complex-specific and 52 species-specific probes, with high specificity and very few cross-reactions. The method therefore allows screening for the presence of multiple species in a single reaction. Results on DNA from cultures were concordant with multi-locus sequence analysis.
In a DNA microarray, probes are attached to a solid surface on microscopic slides. Based on the 18S-ITS1-5.8S region, a DNA microarray following a multiplex PCR was developed distinguishing between 14 fungal pathogens including F. solani and F. oxysporum in blood, BAL and tissue samples [64]. Based on ITS1 and ITS2, a similar array was made for 24 species including the F. solani species complex [65].
Microsphere or liquid phase array analysis combines a multiplex PCR with hybridization to probes linked to fluorescent microspheres, followed by analysis with a flow cytometer (Luminex technology). The newest version of this technique can detect up to 500–1000 different microsphere-linked probes simultaneously. Several assays have been based on the ITS2 region and cover commonly occurring species of genera like Aspergillus, Fusarium and Zygomycetes [66, 67]. Buelow et al. [68] devised an array for the identification of common respiratory fungal pathogens, including Aspergillus fumigatus, Rhizopus microsporus, Scedosporium apiospermum and F. solani based on elongation factor 1-α and other genes. O’Donnell et al. [69] based their microsphere array on the rpb2 gene and used it for discrimination between the Fusarium involved in lens-associated keratitis. Detection limits typically range from 0.1 to 1 ng of DNA, depending on the fungus being tested. Luminex Molecular Diagnostics (Toronto, Canada) has analyte-specific reagents (ASRs) available for different clinically important species including one for Fusarium that has been tested on patient samples [70].
DNA Sequencing
The internal transcribed spacer 1 (ITS1) region has been chosen as a general barcode for fungi [71]. However, this locus does not contain enough variation to distinguish, based on single-gene sequences, between many Fusarium species [9]. Pan-fungal primers based on the ITS1 region have been used to amplify and sequence PCR products to identify to the genus level several invasive species including Fusarium spp. This technique has also been applied in tissue samples [41]. Also, the D1 plus D2 regions of the nuclear large subunit rDNA are not variable enough to distinguish to species level [6, 72]. The β-tubulin gene (β-tub), e.g. [46], and RNA polymerase II second largest subunit rpb2 [69] and elongation factor 1-α (ef-1α) [73] have more discriminatory power.
Multi-locus sequence typing (MLST) is currently seen as the best method for the identification of Fusarium isolates to species level [15•]. Different MLST schemes have been proposed for the different species complexes (Table 1). Especially regions of ef-1α and RNA polymerase II second largest subunit (rpb2) have good discriminatory power and are often used in MLST schemes.
For the identification based on DNA sequences, we rely on blast searches in reference databases and assume that they are sufficiently complete and contain correctly identified and annotated entries. However, 10–20 % of the entries in public repositories like GenBank are estimated to be incorrectly identified to the species level [79]. To limit incorrect entries, two curated databases have been developed for the genus Fusarium: the Fusarium-ID database (http://isolate.fusariumdb.org/ [80]) and the Fusarium MLST database (http://www.cbs.knaw.nl/fusarium/ [9]).
Isothermal Amplifications
Isothermal DNA amplification techniques have been developed that amplify DNA at a single temperature as opposed to PCR, which requires three different temperatures. Isothermal assays are considered to be simple, cost effective and rapid methods for the detection of specific genomic DNA fragments. Loop-mediated isothermal amplification (LAMP) assays have so far been applied to several Fusarium plant pathogens [81, 82]. For LAMP, the detection is based on a set of six primers hybridizing to the target gene and amplification, after which the product can be visualized by a nucleic acid dye, often directly in the reaction tube. Rolling circle amplification (RCA) relies on the formation of a circular DNA molecule when the padlock probe—long oligonucleotides, whose ends are complementary to adjacent target sequences—fits exactly on the target sequence that subsequently can be endlessly transcribed and detected directly in the reaction tube or after gel electrophoresis. Specific RCA probes for the F. oxysporum and F. incarnatum-equiseti species complexes have been developed [83].
Peptide-based Identification Techniques
Besides the galactomannan tests described above, much of the enzyme-linked immunosorbent assay (ELISA) work in Fusarium has been targeting the specific mycotoxins they are known to produce in planta, but we do not know whether these compounds are also produced in patient tissue or whether these molecules, if detected, derive from food intake. Most antibodies targeting Fusarium species themselves are not species-specific and hence could be used to determine a human fusariosis, but no diagnosis to species level can be obtained, e.g. [84–86]. Detection limits for ELISA range from 0.1 to 1 μg of mycelium per millilitre of product tested. Several antibodies and ELISA kits for Fusarium are commercially available, but as far as we know, none are used in hospital laboratories.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is an emerging tool for fast identification and classification of cultured microorganisms based on their protein spectra. Different commercial companies supply MALDI-TOF MS systems. Systems like VITEK (bioMérieux, Marcy l'Etoile, France) or Biotyper (Bruker Daltronics Inc, Billerica, MA) come with their own processing software and spectral database with reference strains.
For bacteria and yeasts, the necessary databases are further advanced than those for filamentous fungi. However, the studies done on Fusarium species are very promising with success rates of identification to the species level of 82–99 % [87–90]. Nevertheless, growth conditions like type and phase (solid/liquid) of media, age of the examined culture, type of sample with or without spores and the treatment of the fungal sample can influence the resulting spectra [91–94]. The limiting part for the level of identification for a given platform seems to be the reliability and level of identification of the strains used for the database production.
The abovementioned studies on Fusarium detection by MALDI-TOF MS analysis were all based on pure cultures. Pan et al. [95] in a study on ocular mycoses reported that with MALDI-TOF MS, the fusarioses could be distinguished from the other etiological agents, while Ananthi et al. [96] showed that human tear protein profiles are influenced by the Fusarium pathogen. Studies on Candida infections in the blood stream suggest that also in blood samples, direct fungal detection may be possible, e.g. [97].
Conclusions
Several clinical features can hint to a fusariosis: For disseminated infections, these are positive galactomannan tests and typical necrotic skin lesions. For keratitis, there is the presence of hyaline hyphae, while for superficial skin infections and onychomycosis, no Fusarium-specific traits are recognized. Especially for deep and disseminated infections, rapid diagnosis is of the utmost importance for timely and adequate treatment. Hence, we see a shift in diagnostic tools applied in the hospital laboratory from classic, morphological determination of the etiological agent that sometimes involves prolonged culturing to obtain all necessary structures for species identification to faster DNA or peptide-based diagnostic tools.
After culturing of Fusarium from a patient, MLST identification is currently seen as the best option for species-level identification [15•]. However, if these specific tools are not available in the laboratory, the use of another molecular and/or peptide-based tool is recommended [15•]. Techniques like microsphere/liquid phase arrays and MALDI-TOF may prove faster in making a species-level diagnosis. Combining available tools will enhance the specificity of identification [98].
Fusarium also contain many other compounds beside DNA, peptides and cell wall components that could be used for future diagnostic purposes. Volatile organic compound profiles detected by selected ion flow tube-mass spectrometry (SIFT-MS) may have diagnostic value [99]. Detection of toxic or non-toxic secondary metabolites produced in cultures may be another option [100, 101]. No doubt new techniques will evolve that will make diagnosis even faster and more reliable.
The main goal of diagnostic tools is better survival and recovery chances for the patient by selecting adequate treatment. As survival rates in invasive infections greatly depend on early diagnosis, tools directly applicable in patient samples will become more and more important. Recent research on differences in antifungal susceptibility between species and isolates, e.g. [5, 13], demonstrates the need of species-level identification in the case of a fusariosis.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Geiser DM, Aoki T, Bacon CW, et al. One fungus, one name: defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology. 2013;103:400–8. This paper defines the phylogenetic cluster of what species in view of this review and other works are to be considered as “Fusarium”. This definition is supported by more than 60 experts.
Leslie JF, Summerell BA. The Fusarium laboratory manual. Oxford: Blackwell Publishing Ltd; 2006.
van Diepeningen AD, Al-Hatmi AMS, Brankovics B, de Hoog GS. Taxonomy and clinical spectra of Fusarium species: where do we stand in 2014? Clin Microbiol Rep. 2014;1(1):10–8. This paper details what species from within the genus Fusarium are capable of causing human disease and provide an overview of the actual symptoms caused.
Nucci M, Varon AG, Garnica M, et al. Increased incidence of invasive fusariosis with cutaneous portal of entry. Brazil Emerg Infect Dis. 2013;19:1567–72.
Dalyan Cilo B, Al-Hatmi ASM, Seyedmousavi S, et al. Emerging Fusarioses at a University Hospital in Turkey: a retrospective study 1995–2014. Eur J Clin Microbiol Infect Dis. 2015. doi:10.1007/s10096-015-2405-y.
O'Donnell K, Sutton DA, Fothergill A, et al. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol. 2008;46(8):2477–90.
O’Donnell K, Gueidan C, Sink S, et al. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet Biol. 2009;46(12):936–48.
O'Donnell K, Sutton DA, Rinaldi MG, Gueidan C, Crous PW, Geiser DM. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J Clin Microbiol. 2009;47(12):3851–61.
O'Donnell K, Sutton DA, Rinaldi. Internet-accessible DNA sequence database for identifying Fusaria from human and animal infections. J Clin Microbiol. 2010;48(10):3708–18.
Lortholary O, Obenga G, Biswas P, et al. International retrospective analysis of 73 cases of invasive fusariosis treated with voriconazole. 2010;54(10):4446–4450.
Migheli Q, Balmas V, et al. Molecular phylogenetic diversity of dermatologic and other human pathogenic fusarial isolates from hospitals in Northern and Central Italy. J Clin Microbiol. 2010;48(4):1076–84.
van Diepeningen AD, Feng P, Ahmed S, Sudhadham M, Bunyaratavej S, de Hoog GS. Spectrum of Fusarium infections in tropical dermatology evidenced by multilocus sequencing typing diagnostics. Mycoses. 2015;58(1):48–57.
Al-Hatmi AM, Bonifaz A, de Hoog G, et al. Keratitis by Fusarium temperatum, a novel opportunist. BMC Infect Dis. 2014;14(1):588.
Nucci M, Anaissie E. Fusarium infections in immunocompromised patients. Clin Microbiol Rev. 2007;20(4):695–704.
Tortorano AM, Richardson M, Roilides E, et al. ESCMID & ECMM joint guidelines on diagnosis and management of hyalohyphomycosis: Fusarium spp, Scedosporium spp, and others. Clin Microbiol Infect. 2014;20 Suppl 3:27–46. The recent guidelines give suggestions for the diagnosis of Fusarium and other hyalohyphomycetes and more importantly recommendation on how to treat them once diagnosed.
Kebabcı N, van Diepeningen AD, Ener B, et al. Fatal breakthrough infection with Fusarium andiyazi: new multi-resistant aetiological agent cross-reacting with Aspergillus galactomannan enzyme immunoassay. Mycoses. 2014;57(4):249–55.
Al-Hatmi AMS, van Diepeningen AD, Curfs-Breuker I, de Hoog GS, Meis JF. Specific antifungal susceptibility profiles of opportunists in the Fusarium fujikuroi complex. J Antimicrobial Chemother. 2015;70(4):1068–71.
Vasantha Ruban V, Geraldine P, Kaliamurthy J, Jesudasan CA, Thomas PA. Keratitis due to Fusarium langsethiae: clinical profile, molecular identification, and susceptibility to antifungals. Mycopathologia. 2015. doi:10.1007/s11046-015-9866-5.
Brasnu E, Bourcier T, Dupas B, et al. In vivo confocal microscopy in fungal keratitis. Br J Ophthamol. 2007;91(5):588–91.
Wang L, Zhang J, Sun S, Zhang Y. In vivo confocal microscopic characteristics of fungal keratitis. Life Sci J. 2008;5(1):51–4.
Vaddavalli PK, Garg P, Sharma S, Sangwan VS, Rao GN, Thomas R. Role of confocal microscopy in the diagnosis of fungal and acanthamoeba keratitis. Ophthalmology. 2011;118(1):29–35.
Agarwal T, Bandivadekar P, Satpathy G, Sharma N, Titiyal JS. Detection of fungal hyphae using smartphone and pocket magnifier: going cellular. Cornea. 2015;34(3):355–7.
Anaissie E, Kantarjian H, Ro J, et al. The emerging role of Fusarium infections in patients with cancer. Medicine (Baltimore). 1988;67(2):77–83.
Bryan CS. Clinical implications of positive blood cultures. Clin Microbiol Rev. 1989;2(4):329–53.
Tortorano AM, Esposto MC, Prigitano A, et al. Cross-reactivity of Fusarium spp. In the Aspergillus galactomannan enzyme-linked immunosorbent assay. J Clin Microbiol. 2012;50(3):1051–3.
Rimek D, Singh J, Kappe R. Cross-reactivity of the PLATELIA CANDIDA antigen detection enzyme immunoassay with fungal antigen extracts. J Clin Microbiol. 2003;41(7):3395–8.
Yoshida M, Obayashi T, Iwama A, et al. Detection of plasma (1→3)-β-d-glucan in patients with Fusarium, Trichosporon, Saccharomyces and Acremonium fungaemias. Med Mycol. 1997;35(5):371–4.
Marom EM, Holmes AM, Bruzzi JF, Truong MT, O'Sullivan PJ, Kontoyiannis DP. Imaging of pulmonary fusariosis in patients with hematologic malignancies. AJR Am J Roentgenol. 2008;190(6):1605–9.
Georgiadou SP, Sipsas NV, Marom EM, Kontoyiannis DP. The diagnostic value of halo and reversed halo signs for invasive mold infections in compromised hosts. Clin Infect Dis. 2011;52(9):1144–55.
Marom EM, Kontoyiannis DP. Imaging studies for diagnosing invasive fungal pneumonia in immunocompromised patients. Curr Opin Infect Dis. 2011;24(4):309–14.
Watts JC, Chandler FW. Fusariosis. In: Connor DH, Chandler FW, Schwartz DA, Manz HJ, Lack EE, editors. Pathology of infectious diseases, vol. 2. 2 1st ed. Hong Kong: Stamford, Appleton & Lange Co; 1997. p. 999–1001.
Bennet JE. Introduction to mycoses. In: Mandell JL, Bennet JE, Dolin R, editors. Principles and practice of infectious diseases. 7. Philadelphia: Churchill Livingstone; 2009. p. 3221–4.
Kaufman L, Standard PG, Jalbert M, Kraft DE. Immunohistologic identification of Aspergillus spp. and other hyaline fungi by using polyclonal fluorescent antibodies. J Clin Microbiol. 1997;35(9):2206–9.
Saito T, Imaizumi M, Kudo K, et al. Disseminated Fusarium infection identified by the immunohistochemical staining in a patient with a refractory leukemia. Tohoku J Exp Med. 1999;187(1):71–7.
Hayden RT, Isotalo PA, Parrett T, et al. In situ hybridization for the differentiation of Aspergillus, Fusarium, and Pseudallescheria species in tissue section. Diagn Mol Pathol. 2003;12(1):21–6.
Montone KT, Livolsi VA, Lanza DC, et al. Rapid In-situ hybridization for dematiaceous fungi using a broad-spectrum oligonucleotide DNA probe. Diagn Mol Pathol. 2011;20(3):180–3.
Okubo Y, Shinozaki M, Wakayama M, et al. Applied gene histopathology: identification of Fusarium species in FFPE tissue sections by in situ hybridization. Methods Mol Biol. 2013;968:141–7.
Shinozaki M, Okubo Y, Sasai D, et al. Identification of Fusarium species in formalin-fixed and paraffin-embedded sections by in situ hybridization using peptide nucleic acid probes. J Clin Microbiol. 2011;49(3):808–13.
Montone KT. Differentiation of Fusarium from Aspergillus species by colorimetric in situ hybridization in formalin-fixed, paraffin-embedded tissue sections using dual fluorogenic-labeled LNA probes. Am J Clin Pathol. 2009;132(6):866–70.
Muñoz-Cadavid C, Rudd S, Zaki SR, et al. Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: comparison of five tissue DNA extraction methods using panfungal PCR. J Clin Microbiol. 2010;48(6):2147–53.
Lau A, Chen S, Sorrell T, et al. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J Clin Microbiol. 2007;45(2):380–5.
Olias P, Jacobsen ID, Gruber AD. Fungal species identification from avian lung specimens by single hypha laser microdissection and PCR product sequencing. Med Mycol. 2011;49(1):56–61.
De Hoog GS, Guarro J, Gené J, Figueras MJ. Atlas of clinical fungi. 3rd ed. Utrecht: CD-ROM, CBS-KNAW Fungal Biodiversity Centre; 2011.
Guarro J, Gené J. Fusarium infections. Criteria for the identification of the responsible species. Mycoses. 1992;35(5–6):109–14.
Summerbell RC, Schroers HJ. Analysis of phylogenetic relationship of Cylindrocarpon lichenicola and Acremonium falciforme to the Fusarium solani species complex and a review of similarities in the spectrum of opportunistic infections caused by these fungi. J Clin Microbiol. 2002;40(8):2866–75.
Azor M, Gené J, Cano J, Manikandan P, Venkatapathy N, Guarro J. Less-frequent Fusarium species of clinical interest: correlation between morphological and molecular identification and antifungal susceptibility. J Clin Microbiol. 2009;47(5):1463–8.
Jurado M, Vázquez C, Patiño B, González-Jaén MT. PCR detection assays for the trichothecene-producing species Fusarium graminearum, Fusarium culmorum, Fusarium poae, Fusarium equiseti and Fusarium sporotrichioides. Syst Appl Microbiol. 2005;28(6):562–8.
Landlinger C, Baskova L, Preuner S, Willinger B, Buchta V, Lion T. Identification of fungal species by fragment length analysis of the internally transcribed spacer 2 region. Eur J Clin Microbiol Infect Dis. 2009;28(6):613–22.
Ahmad S, Khan ZU, Theyyathel AM. Development of a nested PCR assay for the detection of Fusarium solani DNA and its evaluation in the diagnosis of invasive fusariosis using an experimental mouse model. Mycoses. 2010;53(1):40–7.
Sugawara Y, Nakase K, Nakamura A, et al. Clinical utility of a panfungal polymerase chain reaction assay for invasive fungal diseases in patients with haematologic disorders. Eur J Haematol. 2013;90(4):331–9.
Healy M, Reece K, Walton D, et al. Use of the Diversi Lab System for species and strain differentiation of Fusarium species isolates. J Clin Microbiol. 2005;43(10):5278–80.
Lau A, Sorrell TC, Lee O, Stanley K, Halliday C. Colony multiplex-tandem PCR for rapid, accurate identification of fungal cultures. J Clin Microbiol. 2008;46(12):4058–60.
Faria CB, Abe CA, da Silva CN, Tessmann DJ, Barbosa-Tessmann IP. New PCR assays for the identification of Fusarium verticillioides, Fusarium subglutinans, and other species of the Gibberella fujikuroi complex. Int J Mol Sci. 2012;13(1):115–32.
Bernal-Martínez L, Buitrago MJ, Castelli MV, Rodríguez-Tudela JL, Cuenca-Estrella M. Detection of invasive infection caused by Fusarium solani and non-Fusarium solani species using a duplex quantitative PCR-based assay in a murine model of fusariosis. Med Mycol. 2012;50(3):270–5.
Muraosa Y, Schreiber AZ, Trabasso P, et al. Development of cycling probe-based real-time PCR system to detect Fusarium species and Fusarium solani species complex (FSSC). Int J Med Microbiol. 2014;304(3–4):505–11.
Gu Z, Buelow DR, Petraitiene R, Petraitis V, Walsh TJ, Hayden RT. Quantitative multiplexed detection of common pulmonary fungal pathogens by labeled primer polymerase chain reaction. Arch Pathol Lab Med. 2014;138(11):1474–80.
Scauflaire J, Godet M, Gourgue M, Liénard C, Munaut F. A multiplex real-time PCR method using hybridization probes for the detection and the quantification of Fusarium proliferatum, F. subglutinans, F. temperatum, and F. verticillioides. Fungal Biol. 2012;116(10):1073–80.
Baayen RP, O’Donnell K, Waalwijk C, et al. Gene genealogies and AFLP analyses within the Fusarium oxysporum complex identify monophyletic and non-monophyletic formae speciales causing wilt and rot disease. Phytopathology. 2000;90(8):891–900.
O'Donnell K, Sutton DA, Rinaldi MG, et al. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J Clin Microbiol. 2004;42(11):5109–20.
Park JM, Kim GY, Lee SJ, et al. Comparison of RAPD, AFLP, and EF-1α sequences for the phylogenetic analysis of Fusarium oxysporum and its formae speciales in Korea. Mycobiology. 2006;34(2):45–55.
Jureen R, Koh TH, Wang G, et al. Use of multiple methods for genotyping Fusarium during an outbreak of contact lens associated fungal keratitis in Singapore. BMC Infect Dis. 2008;8:92.
Godoy P, Cano J, Gené J, Guarro J, Höfling-Lima AL, Lopes CA. Genotyping of 44 isolates of Fusarium solani, the main agent of fungal keratitis in Brazil. J Clin Microbiol. 2004;42(1):4494–7.
Wang H, Xiao M, Kong F, et al. Accurate and practical identification of 20 Fusarium species by seven-locus sequence analysis and reverse line blot hybridization, and an in vitro antifungal susceptibility study. J Clin Microbiol. 2011;49(5):1890–8.
Spiess B, Seifarth W, Hummel M, et al. DNA microarray-based detection and identification of fungal pathogens in clinical samples from neutropenic patients. J Clin Microbiol. 2007;45(11):3743–53.
Campa D, Tavanti A, Gemignani F, et al. DNA microarray based on arrayed-primer extension technique for identification of pathogenic fungi responsible for invasive and superficial mycoses. J Clin Microbiol. 2008;46(3):909–15.
Landlinger C, Preuner S, Willinger B, et al. Species-specific identification of a wide range of clinically relevant fungal pathogens by use of Luminex xMAP technology. J Clin Microbiol. 2009;474:1063–73.
Liao MH, Lin JF, Li SY. Application of a multiplex suspension array for rapid and simultaneous identification of clinically important mold pathogens. Mol Cell Probes. 2012;26(5):188–93.
Buelow DR, Gu Z, Walsh TJ, Hayden RT. Evaluation of multiplexed PCR and liquid-phase array for identification of respiratory fungal pathogens. Med Mycol. 2012;50(7):775–80.
O’Donnell K, Sarver BAJ, Brandt M, et al. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic Fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreaks of 2005 and 2006. J Clin Microbiol. 2007;45(7):2235–48.
Babady NE, Miranda E, Gilhuley KA. Evaluation of Luminex xTAG fungal analyte-specific reagents for rapid identification of clinically relevant fungi. J Clin Microbiol. 2011;49(11):3777–82.
Schoch CL, Seifert KA, Huhnhof S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A. 2012;109(16):6241–6.
Zhang N, O'Donnell K, Sutton DA, et al. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J Clin Microbiol. 2006;44(6):2186–90.
Knutsen AK, Torp M, Holst-Jensen. Phylogenetic analyses of the Fusarium poae, Fusarium sporotrichioides and Fusarium langsethiae species complex based on partial sequences of the translation elongation factor-1 alpha gene. Int J Food Microbiol. 2004;95(3):287–95.
Schroers HJ, O’Donnell K, Lamprecht S, et al. Taxonomy and phylogeny of the Fusarium dimerum species group. Mycologia. 2009;101(1):44–70.
O’Donnell K, Nirenberg HI, Aoki T, Cigelnik E. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 2000;41:61–78.
Cunnington J. Novel primers developed from mitochondrial intergenic spacers are useful for multi-locus sequence typing of Fusarium oxysporum strains. Eur J Plant Pathol. 2006;116(1):77–80.
Debourgogne A, Gueidan C, Hennequin C, Contet-Audonneau N, de Hoog S, Machouart M. Development of a new MLST scheme for differentiation of Fusarium solani species complex (FSSC) isolates. J Microbiol Methods. 2010;82(3):319–23.
Yli-Mattila T, Paavanen-Huhtala S, Bulat SA, Alekhina IA, Nirenberg HI. Molecular, morphological and phylogenetic analysis of the Fusarium avenaceum/F. arthrosporioides/F. tricinctum species complex—a polyphasic approach. Mycol Res. 2002;106(6):655–69.
Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson KH, Kõljalg U. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One. 2006;1:e59.
Geiser DM, Jimenez-Gasco M, Kang S, et al. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 2004;110:473–9.
Niessen L, Vogel RF. Detection of Fusarium graminearum DNA using a loop-mediated isothermal amplification (LAMP) assay. Int J Food Microbiol. 2010;140(2–3):183–91.
Almasi MA, Dehabadi SMH, Moradi A, Eftekhari Z, Ojaghkandi MA, Aghaei S. Development and application of loop-mediated isothermal amplification assay for rapid detection of Fusarium oxysporum f. sp. lycopersici. J Plant Pathol Microb. 2013;4:5.
Davari M, van Diepeningen AD, Babai-Ahari A, et al. Rapid identification of Fusarium graminearum species complex using rolling circle amplification (RCA). J Microbiol Methods. 2012;89(1):63–70.
Iyer MS, Cousin MA. Immunological detection of Fusarium species in cornmeal. J Food Prot. 2003;66(3):451–6.
Kitagawa T, Sakamoto Y, Furumi K, Ogura H. Novel enzyme immunoassays for specific detection of Fusarium oxysporum f. sp. cucumerinum and for general detection of various Fusarium species. Phytopathology. 1989;79:162–5.
Hill NS, Hiatt EE, Chanh TC. ELISA analysis for Fusarium in barley. Crop Sci. 2006;46:2636–42.
Marinach-Patrice C, Lethuillier A, Marly A, et al. Use of mass spectrometry to identify clinical Fusarium isolates. Clin Microbiol Infect. 2009;15(7):634–42.
De Carolis E, Posteraro B, Lass-Flörl C, et al. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2012;18(5):475–84.
Del Chierico F, Masotti A, Onori M, et al. MALDI-TOF MS proteomic phenotyping of filamentous and other fungi from clinical origin. J Proteom. 2012;75(11):3314–30.
Triest D, Stubbe D, De Cremer K, et al. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of molds of the Fusarium genus. J Clin Microbiol. 2015;53(2):465–76.
Mancini V, Dapporto L, Baracchin D, Luchi N, Turillazzi S, Capretti P. Phenotypic characterization of cryptic Diplodia species by MALDI-TOF MS and the bias of mycelium age. Forest Pathol. 2013;43(6):455–61.
Valentine N, Wunschel S, Wunschel D, Petersen C, Wahl K. Effect of culture conditions on microorganism identification by matrix-assisted laser desorption ionization mass spectrometry. Appl Environ Microbiol. 2005;71(1):58–64.
Reich M, Bosshard PP, Stark M, Beyser K, Borgmann S. Species identification of bacteria and fungi from solid and liquid culture media by MALDI-TOF mass spectrometry. J Bacteriol Parasitol. 2013;S5:002. doi:10.4172/2155-9597.S5-002.
Buskirk AD, Hettick JM, Chipinda I, et al. Fungal pigments inhibit the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis of darkly pigmented fungi. Anal Biochem. 2011;411(1):122–8.
Pan YL, Kuo MT, Chang TC, Ho YP, Chang HC. Characterizing common pathogens of ocular mycoses based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Mycoses. 2012;55(S4):293.
Ananthi S, Venkatesh Prajna N, Lalitha P, Valarnila M, Dharmalingam K. Pathogen induced changes in the protein profile of human tears from Fusarium keratitis patients. PLoS One. 2013;8:e53018.
Marinach-Patrice C, Fekkar A, Atanasova R, et al. Rapid species diagnosis for invasive candidiasis using mass spectrometry. PLoS One. 2010;5:e8862.
Khan ZU, Ahmad S, Theyyathel AM. Diagnostic value of DNA and (1→3)-beta-D-glucan detection in serum and bronchoalveolar lavage of mice experimentally infected with Fusarium oxysporum. J Med Microbiol. 2008;57(Pt1):36–42.
Scotter JM, Langford VS, Wilson PF, McEwan MJ, Chambers ST. Real-time detection of common microbial volatile organic compounds from medically important fungi by selected ion flow tube-mass spectrometry (SIFT-MS). J Microbiol Methods. 2005;63(2):127–34.
Stępień L. The use of Fusarium secondary metabolite biosynthetic genes in chemotypic and phylogenetic studies. Crit Rev Microbiol. 2014;40(2):176–85.
Thrane U. Grouping Fusarium section Discolor isolates by statistical analysis of quantitative high performance liquid chromatographic data on secondary metabolite production. JMicrobiol Meth. 1990;12:23–39.
Acknowledgement
Balázs Brankovics was supported by the Division for Earth and Life Sciences (ALW) with financial aid from the Netherlands Organization for Scientific Research (NWO) (Dossier 833.13.006).
Compliance with Ethics Guidelines
ᅟ
Conflict of Interest
Anne D. van Diepeningen, Jearidienne Iltes, Theo A.J. van der Lee and Cees Waalwijk declare that they have no competing interests.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Advances in Diagnosis of Invasive Fungal Infections
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
van Diepeningen, A.D., Brankovics, B., Iltes, J. et al. Diagnosis of Fusarium Infections: Approaches to Identification by the Clinical Mycology Laboratory. Curr Fungal Infect Rep 9, 135–143 (2015). https://doi.org/10.1007/s12281-015-0225-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12281-015-0225-2