Abstract
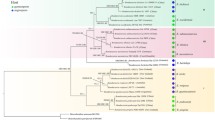
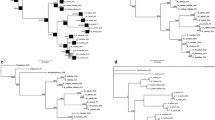
Phylogenetic relationships within theGibberella fujikuroi species complex were extended to newly discovered strains using nucleotide characters obtained by sequencing polymerase chain reaction (PCR) amplified DNA from 4 loci used in a previous study [nuclear large subunit 28S rDNA, nuclear ribosomal internal transcribed spacer (ITS) region, mitochondriaal small subunit (mtSSU) ribosomal DNA, and β-tubulin] together with two newly sampled protein-encoding nuclear genes, translation elongation factor EF-1α and calmodulin. Sequences from the ribosomal ITS region were analyzed separately and found to contain of two highly divergent, nonorthologous ITS2 types. Phylogenetic analysis of the individual and combined datasets identified 10 new phylogenetically distinct species distributed among the following three areas: 2 within Asia and 4 within both Africa and South America. Hypotheses of the monophyly ofFusarium subglutinans and its two formae speciales, f. sp.pini and f. sp.ananas, were strongly rejected by a likelihood analysis. Maximum parsimony results further indicate that the protein-encoding nuclear genes provide considerably more phylogenetic signal that the ribosomal genes sequenced. Relative apparent synapomorphy analysis was used to detect long-branch attraction taxa and to obtain a statistical measure of phylogenetic signal in the individual and combined datasets.
Similar content being viewed by others
Literature cited
Aoki, T. and O’Donnell, K. 1999. Morphological and molecular characterization ofFusarium pseudograminearum sp. nov., formerly recognized as the Group 1 population ofF. graminearum. Mycologia91: 597–609.
Aoki, T. and O’Donnell, K. 1999b. Morphological characterization ofGibberella coronicola sp. nov., obtained through mating experiments ofFusarium pseudograminearum. Mycoscience40: 443–453.
Appel, D. J. and Gordon, T. R. 1996. Relationships among pathogenic and nonpathogenic isolates ofFusarium oxysporum based on the partial sequence of the intergenic spacer region of the ribosomal DNA. Mol. Pl.-Microbe Interact.9: 125–138.
Bain, L. J. and Engelhardt, M. 1992. Introduction to probability and mathematical statistics. Duxbury Press, Belmont, California.
Baldwin, B. G., Sanderson, M. J., Porter, J. M., Wojciechowski, M. F., Campbell, C. S. and Donoghue, M. J. 1995. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Missouri Bot. Gard.82: 247–277.
Bolkan, H. A., Dianese, J. C. and Cupertino, F. P. 1979. Pineapple flowers as principal infection sites forFusarium moniliforme var.subglutinans. Pl. Dis. Reptr.63: 655–657.
Booth, C. 1971. The genusFusarium. CMI, Kew, Surrey.
Britz, H., Coutinho, T. A., Wingfield, M. J., Marasas, W. F. O., Gordon, T. R. and Leslie, J. F. 1999.Fusarium subglutinans f. sp.pini represents a distinct mating population in theGibberella fujikuroi species complex. Appl. Environm. Microbiol.65: 1198–1201.
Bruns, T. D., Szaro, T. M., Gardes, M., Cullings, K. W., Pan, J. J., Taylor, D. L., Horton, T. R., Kretzer, A., Garbelotton, M. and Li, Y. 1998. A sequence database for the identification of ectomycorrhizal basidiomycetes by phylogenetic analysis. Mol. Ecol.7: 257–272.
Buckler, E. S. and Holtsford, T. P. 1996.Zea systematics: Ribosomal ITS evidence. Mol. Biol. Evol.13: 612–622.
Carbone, I., Anderson, J. B. and Kohn, L. M. 1999, Patterns of descent in clonal lineages and their multilocus fingerprints are resolved with combined gene genealogies. Evolution53: 11–21.
Cardá-Olmedo, E., Fernández-Martín, R. and Ávalos, J. 1994. Genetics and gibberellin production inGibberella fujikuroi. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol.65: 217–225.
Correll, J. C., Gordon, T. R. and McCain, A. H. 1992. Genetic diversity in California and Florida populations of the pitch canker fungusFusarium subglutinans f. sp.pini. Phytopathology82: 415–420.
Cunningham, C. W. 1997. Can three incongruence tests predict when data should be combined? Mol. Biol. Evol.14: 733–740.
Ellis, J. J. 1988. Lesser known species in sectionLiseola ofFusarium and near relatives in theFusarium oxysporum group. Mycologia80: 734–738.
Freeman, S., Maimon, M. and Pinkas, Y. 1999. Use of GUS transformants ofFusarium subglutinans for determining etiology of mango malformation disease. Phytopathology89: 456–461.
Graham, S. W., Kohn, J. R., Morton, B. R., Eckenwalder, J. E. and Barrett, S. C. H. 1998. Phylogenetic congruence and discordance among one morphological and three molecular data sets from Pontederiaceae.. Syst. Biol.47: 545–567.
Gardes, M. and Bruns, T. D. 1993. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol.2: 112–118.
Geiser, D. M., Pitt, J. I. and Taylor, J. W. 1998. Cryptic speciation and recombination in the aflatoxin-producing fungusAspergillus flavus. Proc. Natl. Acad. Sci. USA95: 388–393.
Gerlach, W. and Nirenberg, H. 1982. The genusFusarium—A pictorial atlas. Mitt. Biol. Bundesanst. Land- u. Forstwirtsch. Berlin-Dahlem209: 1–406.
Hillis, D. M. and Bull, J. J. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol.42: 182–192.
Kellogg, E. A., Appels, R. and Mason-Gamer, R. J. 1996. When genes tell different stories: the diploid genera of Triticeae (Gramineae). Syst. Bot.21: 321–347.
Kluge, A. G. 1988. Parsimony in vicariance biogeography: a quantitative method and a greater antillean example. Syst. Zool.37: 315–328.
Kumar, J., Singh, U. S. and Beniwal, S. P. S. 1993. Mango malformation: one hundred years of research. Annu. Rev. Phytopathol.31: 217–232.
Leslie, J. E. 1995.Gibberella fujikuroi: available populations and variable traits. Can. J. Bot.73: S282-S291.
Lutzoni, F. 1997. Phylogeny of lichen-and non-lichen-forming omphalinoid mushrooms and the utility of testing for combinability among multiple data sets. Syst. Biol.46: 373–406.
Lutzoni, F. and Vilgalys, R. 1995. Integration of morphological and molecular data sets in estimating fungal phylogenies. Can. J. Bot.73: S649-S659.
Lyons-Weiler, J. 1999. RASA 2.3 software and documentation for the MAC. http:test 1.bio.psu.edu/LW/rasatext/html.
Lyons-Weiler, J. and Hoelzer, G. A. 1997. Escaping from the Felsenstein zone by detecting long branches in phylogenetic data. Mol. Phylo. Evol.8: 375–384.
Lyons-Weiler, J., Hoelzer, G. A. and Tausch, R. J. 1996. Relative apparent synapomorphy analysis (RASA) I: The statistical measurement of phylogenetic signal. Mol., Biol. Evol.13: 749–757.
Maddison, W. P. and Maddison, D. R. 1992. MacClade: Analysis of phylogeny and character evolution, Version 3. Sinauer Associates, Sunderland, MA.
Matuo, T. and Snyder, W. C. 1973. Use of morphology and mating populations in the identification of formae speciales inFusarium solani. Phytopathology63: 562–565.
Moretti, A. A., Logrieco, A., Bottalico, A., Ritieni, A., Fogliano, V. and Randazzo, G. 1996. Diversity in beauvericin and fusaproliferin production by different populations ofGibberella fujikuroi (Fusarium sectionLiseola). Sydowia48: 44–56.
Nelson, P. E., Plattner, R. D., Shackelford, D. D. and Desjardins A. E. 1992. Fumonisin B1 production byFusarium species other thanF. moniliforme in sectionLiseola and by some related species. Appl. Environm. Microbiol.58: 984–989.
Nelson, P. E., Toussoun, T. A. and Marasas, W. F. O. 1983.Fusarium Species: An illustrated manual for identification. Pennsylvania State University, University Park, PA.
Nirenberg, H. 1976. Untersuchungen über die morphologische und biologische Differenzierung in derFusarium-SektionLiseola. Mitt. Biol. Bundesanst. Land- u. Forstwirtsch. Berlin-Dahlem169: 1–117.
Nirenberg, H. and O’Donnell, K. 1998. NewFusarium species and combinations within theGibberella fujikuroi species complex. Mycologia90: 434–458.
Nirenberg, H., O’Donnell, K., Kroschel, J., Andrianaivo, A. P., Frank, J. M. and Mubatanhema, W. 1998. Two new species ofFusarium: Fusarium brevicatenulatum from the noxious weedStriga asiatica in Madagascar andFusarium pseudoanthophilum fromZea mays in Zimbabwe. Mycologia90: 459–464.
O’Donnell, K. 1998. Molecular genetic approaches for measuring microbial diversity: Examples from filamentous fungi. In: Diversity and use of agricultural microorganisms: The fifth international workshop on genetic resources, (ed. by National Institute of Agrobiological Resources), pp. 7–24. National Institute of Agrobiological Resources, Tsukuba, Japan.
O’Donnell, K. and Cigelnik, E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungusFusarium are nonorthologous. Mol. Phylo. Evol.7: 103–116.
O’Donnell, K., Cigelnik, E. and Nirenberg, H. I. 1998a. Molecular systematics and phylogeography of theGibberella fujikuroi species complex. Mycologia90: 465–493.
O’Donnell, K. and Gray, L. E. 1995. Phylogenetic relationships of the soybean sudden death syndrome pathogenFusarium solani f. sp.phaseoli inferred from rDNA sequence data and PCR primers for its identification. Mol. Pl. Microbe Interact.8: 709–716.
O’Donnell, K., Kistler, H. C., Cigelnik, E. and Ploetz, R. C. 1998b. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA95: 2044–2049.
Rai, B. and Upadhyay, R. S. 1982.Gibberella indica: the perfect state ofFusarium udum. Mycologia74: 343–346.
Sanders, I. R., Alt, M., Groppe, K., Boller, T. and Wiemken, A. 1995. Identification of ribosomal DNA polymorphisms among and within spores of the Glomales: application to studies on the genetic diversity of arbuscular mycorrhizal fungal communities. New Phytol.130: 419–427.
Schardl, C. L., Leuchtmann, A., Tsai, H.-F., Collett, M. A., Watt, D. M. and Scott, D. B. 1994. Origin of a fungal sumbiont of perennial ryegrass by interspecific hybridization of a mutualist with the ryegrass choke pathogen,Epichloë typhina. Genetics136: 1307–1317.
Seifert, K. A. and Louie-Seize, G. 1999. Phylogeny and species concepts in thePenicillium aurantiogriseum complex as inferred from partial beta-tubulin gene DNA sequences. Proceedings 3rd International Workshop onPenicillium andAspergillus, Baarn, The Netherlands.
Simpson, B. B. and Ogorzaly, M. C. 1995. Economic botany: Plants in our world, McGraw-Hill, New York.
Sorenson, M. D. 1996. TreeRot. Computer program and documentation. University of Michigan. Ann Arbor, MI.
Steenkamp, E. T., Wingfield, B. D., Coutinho, T. A., Wingfield, M. J. and Marasas, W. F. O. 1999. Differentiation ofFusarium subglutians f. sp.pini by histone gene sequence data. Appl. Environm. Microbiol.65: 3401–3406.
Sun, S.-K. and Snyder, W. C. 1981. The bakanae disease of the rice plant. In:Fusarium: Diseases, biology, and taxonomy, (ed. by Nelson, P. E., Toussoun, T. A. and Cook, R. J.), pp. 104–113. Pennsylvania State University. University Park, PA.
Swofford, D. L. 1998. PAUP*4.0: Phylogenetic analysis using parsimony. Sinauer Associates. Sunderland, MA.
Templeton, A. 1983. Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution37: 221–244.
Ventura, J. A. 1998. Resistance ofFusarium subglutinans f. sp.ananas to benomyl. 8th Intern.Fusarium Workshop, Egham, England, August 17–20.
Viljoen, A., Marasas, W. F. O., Wingfield, M. J. and Viljoen, C. D. 1997. Characterization ofFusarium subglutinans f. sp.pini causing root disease ofPinus patula seedlings in South Africa. Mycol. Res.101: 437–445.
Volger, A. P. and DeSalle, R. 1994. Evolution and phylogenetic information content of the ITS-1 region in the tiger beetleCicindela dorsalis. Mol. Biol. Evol.11: 393–405.
Waalwijk, C., De Koning, J. R. A., Baayen, R. P. and Gams, W. 1996. Discordant groupings ofFusarium spp. from sectionsElegans, Liseola andDlaminia based on ribosomal ITS1 and ITS2 sequences. Mycologia88: 316–328.
Wendel, J. F. and Doyle, J. J. 1998. Phylogenetic incongruence: Window into genome history and molecular evolution. In: Molecular systematics of plants, (ed. by Soltis, P. S., Soltis, D. E. and Doyle, J. J.), pp. 265–296. Chapman and Hall, New York.
Wendel, J. F., Schnabel, A. and Seelanan, T. 1995. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl. Acad. Sci. USA92: 280–284.
White, T. J., Bruns, T., Lee, S. and Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: A guide to methods and application, (ed. by Innis, M. A., Gelfand, D. H., Sninsky, J. J. and White, T. J.), pp. 315–322. Academic Press, New York.
Wollenweber, H. W. and Reinking, O. A. 1935. Die Fusarien ihre Beschreibung, Schadwirkung und Bekämpfung. Paul Parey, Berlin.
Zijlstra, C., Lever, A. E. M., Uenk, B. J. and Van Silfhout, C. H. 1995. Differences between ITS regions of isolates of rootknot nematodesMeloidogyne hapla andM. chitwoodi. Phytopathology85: 1231–1237.
Author information
Authors and Affiliations
About this article
Cite this article
O’Donnell, K., Nirenberg, H.I., Aoki, T. et al. A multigene phylogeny of theGibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 41, 61–78 (2000). https://doi.org/10.1007/BF02464387
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02464387