Abstract
Hymenochaetales Oberw. is an order classified in Basidiomycota of Fungi, and species in this order display notable diversity. They exhibit various fruiting body shapes, including clavarioid, effused-reflexed, and resupinate basidiomes. Few mycorrhizal species have been reported in Hymenochaetales, but wood-decaying species dominate the order. Hymenochaetaceae Imazeki & Toki and Schizoporaceae Jülich are the most species-rich families within Hymenochaetales, and most species in the Republic of Korea belong to these two families. As such, current taxonomic classification and nomenclature are not reflected upon species in the remaining Hymenochaetales families. For this study, a multifaceted morphological and multigenetic marker-based phylogenetic investigation was conducted to, firstly, comprehensively identify understudied Hymenochaetales specimens in Korea and, secondly, reflect the updates on the species classification. Five genetic markers were assessed for the phylogenetic analysis: nuclear small subunit ribosomal DNA (nSSU), internal transcribed spacer (ITS), nuclear large subunit ribosomal DNA (nLSU), RNA polymerase II subunit 2 gene (RPB2), and translation elongation factor 1 gene (TEF1). The results from phylogenetic analysis supported 18 species classified under eight families (excluding Hymenochaetaceae and Schizoporaceae) in Korea. Species formerly placed in Rickenellaceae and Trichaptum sensu lato have been systematically revised based on recent taxonomic reconstructions. In addition, our findings revealed one new species, Rickenella umbelliformis, and identified five formerly nationally unreported species classified under five understudied families. Our findings contribute to a better understanding of Hymenochaetales diversity and highlight the need for continued research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hymenochaetales Oberw. was once considered a group of fungi causing white rot, bearing setae, and having an imperforate parenthosome and no clamps (Hibbett & Thorn, 2001). However, multiple taxonomic revisions based on nuclear large subunit ribosomal DNA (nLSU) phylogeny (Larsson, 2007; Redhead et al., 2002) have introduced external basidiomycetes into Hymenochaetales that cumulated exceptions to these commonalities (van Driel et al., 2009). Now, the order includes mycorrhizal species, such as Coltricia species (Tedersoo et al., 2007), and species of diverse morphological characters. Various basidiome types are found across the order, such as clavarioid, effused-reflexed, resupinate, stipitate stereoid, and stipitate lamellate basidiomes (Dentinger & McLaughlin, 2006; Larsson et al., 2006). Collectively, 69 Hymenochaetales genera have been reported up to date (Wang et al., 2023; Zhou et al., 2023), with species numbers growing rapidly since the last estimate of 600 in 2008 (Kirk et al., 2008).
Most Hymenochaetales species are classified under two well-known families, Hymenochaetaceae Imazeki & Toki and Schizoporaceae Jülich, from 16 accepted families. Many families of the remaining 14 are small in species numbers, several being monogeneric. Most have been recently supported at a family level based on a concatenated dataset of multiple genetic markers, including nuclear small subunit ribosomal DNA (nSSU), internal transcribed spacer (ITS), nLSU, mitoribosomal small subunit (mt-SSU), translation elongation factor 1 gene (TEF1), RNA polymerase II subunit 1 gene (RPB1), and RNA polymerase II subunit 2 gene (RPB2) (Wang et al., 2023; Zhou et al., 2023). Compared to the traditional use of ITS and nLSU markers, multiple genetic markers have significantly increased the resolution of taxon delimitation in Hymenochaetales, revealing numerous new species and allowing better species identification (Cho et al., 2023; Liu et al., 2021).
Reports of indigenous Hymenochaetales species in the Republic of Korea were also concentrated within Hymenochaetaceae and Schizoporaceae (Jang et al., 2016; Kim et al., 2009, 2016). The incorporation of molecular data in evaluating specimens from Hymenochaetaceae and Schizoporaceae has expanded the documentation of species previously unreported within the country (Cho et al., 2021; Kim et al., 2016), and this has contributed to a more comprehensive account of species diversity in these families. For the others, there were only two families (Rickenellaceae Vizzini and Tubulicrinaceae Jülich), six genera, and fourteen species registered in the national list (https://species.nibr.go.kr/; accessed 2023-11-15), where most were identified based on macromophological characters and/or a single genetic marker (Kim et al., 1976; Park et al., 2017). A more profound research of these families based on multigenetic marker phylogeny would accurately identify or verify these species and unveil the true diversity of Hymenochaetales in Korea.
The main objectives of the current study were to (i) identify species of understudied Hymenochaetales families in Korea and (ii) reflect the latest systematic classification for these species. To achieve these two objectives, we conducted a multifaceted morphological observation and multigenetic marker-based phylogenetic analysis. Five genetic markers were analyzed for phylogeny, namely nSSU, ITS, nLSU, RPB2, and TEF1. These regions had been selected based on the ease of amplification, thus acquisition of sequences, and the availability of reference sequences in the public database. As a result, one new species was confirmed and described, and the genus Sidera and five species, each in a different family, were revealed for the first time in Korea.
Materials and Methods
Study Materials
Hymenochaetales specimens collected from Korea were stored as dry specimens at the Seoul National University Fungus Collection (SFC). Twenty-one specimens classified under understudied Hymenochaetales families (excluding Hymenochaetaceae and Schizoporaceae) were selected for the present study based on preliminary ITS-based species identification (methods described below). The selected specimens were collected over the period of 2014 to 2023. Field photographs and information on the collection date, location, and additional notes of fresh fruiting bodies were available for each specimen.
DNA Extraction, PCR, and Sequencing
Fungal tissue of approximately 1 × 1 cm was isolated from each specimen for genomic DNA extraction. Tissue grinding was done using a Bead Ruptor Elite (OMNI International) in 200 μl of 2 × Cetyltrimethyl ammonium bromide (CTAB) buffer. The remaining steps were conducted using the AccuPrep Genomic DNA Extraction Kit (Bioneer).
For the polymerase chain reaction (PCR), a PCR reaction mixture was prepared using a PCR Premix (Bioneer). For each reaction, 1–2 µl of DNA was used. PCR was performed using a C1000 thermal cycler (Bio-Rad). The ITS region was amplified with ITS1F and ITS4B primers (Gardes & Bruns, 1993) under the following conditions: 95℃ for 5 min; 35 cycles of 95℃ for 40 s, 55℃ for 40 s, and 72℃ for 1 min; and 72℃ for 5 min. The nLSU region was amplified with LR0R and LR5/LR7 primers (Vilgalys & Hester, 1990) under the following conditions: 95℃ for 5 min; 35 cycles of 95℃ for 40 s, 55℃ for 40 s, and 72℃ for 1.5 min; and 72℃ for 5 min. The nSSU region was amplified with NS1 and NS4 primers (White et al., 1990) under the same condition for nLSU. The RPB2 region was amplified with fRPB2-5F and fRPB2-7.1R primers (Matheny, 2005) under the following conditions: 95℃ for 5 min; 10 cycles of 95℃ for 40 s, 60℃ for 40 s, and 72℃ for 2 min; 35 cycles of 95℃ for 40 s, 55℃ for 1.5 min, 72℃ for 2 min; and 72℃ for 10 min. The TEF1 region was amplified with EF1-983F and EF1-1567R primers (Rehner & Buckley, 2005) under the following conditions: 95℃ for 5 min; 35 cycles of 95℃ for 40 s, 59℃ for 40 s, and 72℃ for 1.5 min; and 72℃ for 5 min.
All PCR products were verified by gel electrophoresis using a 1% agarose gel and Gel Doc™ XR (Bio-Rad). The PCR products were purified using the Expin™ PCR Purification Kit (GeneAll Biotechnology). DNA sequencing was performed with the PCR primers at Bioneer using an ABI Prism 3730XL machine (Applied Biosystems). Preliminary identification for sequences was performed through NCBI BLAST (using the blastn database) to eliminate contaminated data and select specimens of understudied Hymenochaetales families based on ITS. After a manual quality check for chimeras, mixed peaks, and noise, the forward and reverse sequence reads were assembled using Geneious Prime 2023.0.3 (http://www.geneious.com/). The final sequences are deposited in GenBank (Table 1).
Phylogenetic Analyses
The newly generated sequences were aligned for each genetic region using MAFFT version 7 (Katoh & Standley, 2013) with reference sequences from GenBank, including those derived from specimens in Korea and other type-derived or published Hymenochaetales sequences (Table 1). Manual trimming was performed at the ends of the alignments. Through a preliminary maximum likelihood (ML) phylogenetic tree inference for each genetic marker, any sequence regarded as non-Hymenochaetales (e.g., MK269149 & OL462828) was removed for the final tree inference. The five genetic regions were concatenated using Geneious Prime 2023.0.3.
ML and Bayesian inference (BI) trees were constructed using the concatenated dataset, and Fomitopsis pinicola AFTOL 770, Heterobasidion annosum 06129/6, Polyporus squamosus Cui 10595, and Tremellodendron pallidum AFTOL 699 were used as outgroup (Wang et al., 2023). A nucleotide substitution model for each genetic marker was estimated and employed by respective phylogeny tools. The ML tree was inferred using RAxML v. 8.2.12 (Stamatakis, 2014) with 1,000 replications, and the BI tree was constructed with MrBayes v.3.2.6 (Huelsenbeck & Ronquist, 2001), starting from random trees. BI trees were sampled every 500th generation from one million generations. A 75% support majority rule consensus tree was constructed after removing the first 5% of the trees, and the Bayesian Posterior Probabilities (BPP) were calculated from the remaining trees. Species identification for each specimen was verified based on the final phylogenetic tree.
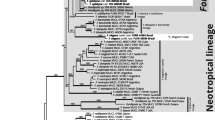
The same inference tools were used to construct Rickenella phylogenetic trees using a concatenated ITS and nLSU dataset. Sequences of all Rickenella specimens were retrieved (2023-10-14) from GenBank as a reference. Outgroup sequences selected were Cantharellopsis prescotii ANT285-HRL2135 and H6059300 and Cotylidia fibrae BJTC FM639 based on their close phylogenetic relationship with Rickenella, as verified in Zhang et al. (2018).
Morphological Analyses
Morphological characters were analyzed for the studied fungarium specimens. Descriptions of macromorphological characters, such as the color, texture, and size of basidiomes, were based on field notes and photographs. For the micromorphological characters, the pileus, hymenophore, and stipe tissues were observed. The tissues were mounted in 5% KOH and stained in 1% Congo red dye. Microscopic features such as basidiospores, basidia, cystidia, and hyphae were observed under a Nikon 80i compound light microscope (Nikon) at 100 × to 400 × magnification.
For the measurements of microscopic features, at least 20 elements of basidia, basidiospores, and cystidia were selected, where possible. The 5% extreme values from each end are given in parentheses. The abbreviation ‘L’ and ‘W’ refer to the mean basidiospore length and width, respectively, ‘Q’ refers to the average length-to-width ratio of basidiospores, and [n/m/p] refers to measurements of n basidiospores from m basidiomes of p collections. Cyanophilic and iodine reactions of basidiospores were tested using Cotton Blue and Melzer’s reagent. All color descriptions follow the codes in the Methuen Handbook of Colours (Kornerup & Wanscher, 1978).
Results
Phylogenetic Analyses
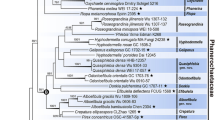
Three to five genetic marker sequences were newly acquired from 15 specimens. The multigenetic marker phylogenetic analysis was conducted using the concatenated alignments of 202 specimen data with 5788 bases, including gaps (Supplementary data 1): nSSU = 1087 bases; ITS = 1499 bases (ITS 1 = 821 bases, 5.8S = 160 bases, ITS2 = 518 bases); nLSU = 1412 bases; RPB2 = 1193 bases (exon 1 = 1106 bases, intron 1 = 62 bases, exon 2 = 25 bases); TEF1 = 597 bases (exon 1 = 150 bases, intron 1 = 85 bases, exon 2 = 107 bases, intron 2 = 89 bases, exon 3 = 166 bases). The BI phylogenetic tree (Fig. S1) topology conformed with the ML tree, supporting all 16 families as independent (Fig. 1).
Phylogeny of Hymenochaetales. The phylogenetic tree was inferred using ML and BI methods (tree topology from ML) based on concatenated nSSU + ITS + nLSU + RPB2 + TEF1 dataset. Statistical values (ML/BI) above 75/0.75 are designated on or below each branch. Strains not indicated by any family are classified under incertae sedis. Colored boxes feature families with species present in Korea, and colored specimens indicate those from Korea
Exclusive of Hymenochaetaceae and Schizoporaceae, 18 species in eight understudied Hymenochaetales families were phylogenetically confirmed to be present in Korea (Fig. 1). Five unreported species were as follows: Rickenella indica in Rickenellaceae, Rigidoporus ginkgonis in Rigidoporaceae, Sidera tibetica in Sideraceae, Skvortzovia dabieshanensis in Skvortzoviaceae, and Tubulicrinis calothrix in Tubulicrinaceae. One species in the Rickenellaceae was phylogenetically revealed to be new to science. The new species, introduced here as Rickenella umbelliformis, was strongly supported (ML/BI support values = 99/1 in Fig. 1), even with two genetic rDNA markers (ITS and nLSU) dataset (85/0.99 in Fig. 2). The remaining 12 indigenous species in Korea were Hirschioporus abietinus, H. acontextus, H. pubescens, and Pallidohirschioporus biformis in Hirschioporaceae, Peniophorella odontiiformis, P. pubera, and P. subpraetermissa in Peniophorellaceae, Resinicium rimulosum in Resiniciaceae, Rickenella fibula in Rickenellaceae, Rigidoporus corticola and R. piceicola in Rigidoporaceae, and Skvortzovia pinicola in Skvortzoviaceae.
Morphological Characters of Native Specimens
A combination of micromorphological characters of Rickenella umbelliformis sp. nov. was distinct from its closely related species, supporting its novelty. The morphology of five nationally unreported species was generally uniform with descriptions of each respective type or a widely accepted specimen. However, some differences were found in the measurements of microscopic features, such as basidiospores and basidia. Detailed comparisons are listed in the Notes sections of Taxonomy.
Taxonomy
This section includes species descriptions of one new species and five previously unreported species from Korea.
Rickenellaceae Vizzini
Rickenella Raithelh.
Rickenella indica K. P. D. Latha & Manim., Fig. 3
Rickenella indica from the Republic of Korea. A Basidiomes of SFC20140626-39. B Micromorphological characters, where ‘s’ refers to basidiospores, ‘b1’ for basidia, ‘b2’ for basidioles, ‘c1’ for cheilocystidia and pleurocystidia, ‘c2’ for pileocystidia, ‘c3’ for caulocystidia, ‘h1’ for hymenial hyphae, ‘h2’ for pileipellis hyphae, and ‘h3’ for stipitipellis hyphae
MycoBank: MB 807702
Description: Basidiomes small, omphalinoid. Pileus 1.0–7.3 mm wide, downy, hygrophanous, paraboloid to convex with a shallow central depression, margin wavy, decurved with age, center reddish orange (9B8), surface yellowish orange (7A8), bleaching towards margin. Stipe 8.0–46.3 × 0.5–1.7 mm, central, cylindrical, downy, gelatinous, brownish yellow (5B7), gradually becomes lighter in color to the base, white at the base.
Basidiospores (5.6–)6.0–6.7(–7.2) × 2.8–3.4(–3.6) µm, L = 6.6 µm, W = 3.1 µm, Q = 1.8–2.2, subcylindrical, thin-walled, smooth, guttules present, cyanophilous, and neither amyloid nor dextrinoid. Basidia (15.5–)16.2–20.4(–20.6) × (4.3–)4.7–5.1 µm, thin-walled, clavate to narrowly utriform, 4-spored. Cheilocystidia and pleurocystidia indistinguishable, 38.8–53.8(–54.6) × (6.5–)7.0–12.0 µm, abundant, narrowly lageniform and obclavate, thin-walled, hyaline. Pileus trama hyphae septate, subregular, cylindrical or subfusoid, 13.3 µm wide on average, moderately thick-walled, hyaline, inamyloid. Pileocystidia 51.9–66.9 × 10.8–16.6 µm, narrowly lageniform to tibiiform, some septate, moderately thick-walled. Stipitipellis hyphae septate, moderately thick-walled, hyaline, inamyloid. Caulocystidia similar to cheilocystidia and pleurocystidia in all aspects. Clamp connections present.
Ecology/Substrate/Host: Grows on moss bed on ground.
Distribution: India, Republic of Korea
Specimens examined: Republic of Korea: Gyeonggi-do, Guri-si, Donggu-dong, Donggureung, deciduous forest, on ground moss bed of Plagiomnium acutum; 26 June 2014, Young Woon Lim (SFC20140626-39). Republic of Korea: Seoul, Seongdong-gu, Seoulsup 2-gil; 37°32′44″N 127°2′19″E, 16 m, Seoul forest park, mixed trees, on ground moss bed of Trachycystis microphylla; 02 July 2023, Shinnam Yoo (SFC20230702-01).
Notes: Rickenella indica from Korea largely resembles the holotype from India morphologically with overlapping ranges of the pileus, basidia, and pleurocystidia in size and also, ecologically, sharing the same lineage of symbiotic moss (Bryopsida). These biogeographically discrete specimens are also supported as the same species in the multigenetic marker phylogeny. However, they are different in several other morphological characters. Rickenella indica from Korea has longer and thicker stipe, more cylindrical basidiospores, and larger pileocystidia than those of the holotype with stipe 3–22 × 0.5–1 µm, basidiospores Q = 1.14–2.16, and pileocystidia similar to cheilocystidia (Latha et al., 2015).
Rickenella umbelliformis Y. Cho & Y.W. Lim, sp. nov. Fig. 4
Rickenella umbelliformis sp. nov. A Basidiomes of SFC20150701-65 (holotype). B Micromorphological characters, where ‘s’ refers to basidiospores, ‘b1’ for basidia, ‘b2’ for basidioles, ‘c1’ for cheilocystidia and pleurocystidia, ‘c2’ for pileocystidia, ‘c3’ for caulocystidia, ‘h1’ for hymenial hyphae, ‘h2’ for pileipellis hyphae, and ‘h3’ for stipitipellis hyphae
MycoBank: MB 849306
Etymology: ‘umbelliformis’ refers to the umbrella shape of the mushroom in Latin.
Holotype: Republic of Korea: Jeju-do, Jeju-si, Jocheon-eup; Jeju Provincial Park entrance Gotjawal, mixed forest, on moss bed of Brachythecium populeum; 1 July 2015, Young Woon Lim (SFC20150701-65, dried specimen).
Description: Basidiomes small, omphalinoid. Pileus 5–15 mm wide, hygrophanous, downy, paraboloid to hemispherical, slightly campanulate when young, forming central depression and wavy decurved margin with age, plicate, center reddish orange (10B8), surface yellowish orange (6A8), bleaching towards margin. Lamellae 12–16, deeply decurrent, white, single lamellula present between two lamellae, edge entire. Stipe 21–42 × 1.2–1.7 mm, central, cylindrical, downy, gelatinous, yellow (4A8) when young, yellowish orange (6A8) when mature, gradually becomes lighter in color to the base, white at the base.
Basidiospores [20/3/2] (4.9–)5.3–6.1(–6.5) × 2.4–2.9(–3.2) µm, L = 5.5 µm, W = 2.8 µm, Q = 1.7–2.2, subcylindrical, thin-walled, smooth, guttules present, cyanophilous, and neither amyloid nor dextrinoid. Basidia (15.9–)16.4–20.0 × (3.3–)3.5–5.4(–5.8) µm, thin-walled, narrowly utriform, 4-spored, sterigmata up to 3.6 µm. Lamellar trama hyphae septate, subregular, cylindrical or subfusoid, moderately thick-walled, hyaline, inamyloid. Cheilocystidia and pleurocystidia indistinguishable, (21.9–)31.6–43.3(–48.0) × (6.1–)6.3–8.9(–9.6) µm, either rare or very frequent, narrowly lageniform and sub-capitate with apex of 2.3–3.6 µm in width, thin-walled, hyaline. Pileipellis hyphae septate, subregular, cylindrical or subfusoid, up to 18.8 µm in width, moderately thick-walled, hyaline, inamyloid, longer than tramal hyphae. Pileocystidia 39.0–48.0(–60.8) × (8.4–)11.4–16.1(–17.8) µm, narrowly lageniform and sub-capitate, moderately thick-walled but thinning towards the apex of 3.0–5.5 µm in width. Stipitipellis hyphae septate, moderately thick-walled, hyaline, inamyloid. Caulocystidia (45.0–)48.2–73.2(– 95.6) × (9.2–)9.5–23.6(– 25.7) µm, narrowly lageniform and sub-capitate, moderately thick-walled but thinning towards the apex of 2.7–4.1 µm in width. Clamp connections observed in lamellar trama only.
Ecology/Substrate/Host: Grows on various Bryophyta moss beds on ground.
Distribution: The Republic of Korea
Additional specimens examined: Republic of Korea: Gyeonggi-do, Guri-si, Donggu-dong, Donggureung, deciduous forest, on ground moss bed of Plagiomnium acutum; 12 May 2016, Hae Jin Cho, Seobihn Lee, Vladimir Li (SFC20160512-10). Republic of Korea: Gyeongsangbuk-do, Ulleung-gun, Ulleung-eup, deciduous forest, on ground moss bed of Trachycystis microphylla; 13 July 2016, Jae young Park, Myung Soo Park, Nam Kyu Kim (SFC20160713-77). Republic of Korea: Jeollanam-do, Wando-gun, Gunoe-myeon, Chopyeong 1-gil, Wando Arboretum, mixed forest, on ground moss bed of Trachycystis microphylla; 4 July 2018, Hae Jin Cho, Ki Hyung Park (SFC20180704-81). Republic of Korea: Seoul, Gwanak-gu, Gwanak-ro; 37°27′34″N 126°56′53″E, 87 m, on ground moss bed of Brachythecium populeum; 12 June 2023, Yoonhee Cho, Dohye Kim, Minseo Cho (SFC20230612-01).
Notes: Rickenella umbelliformis sp. nov. either had very few or abundant cystidia based on different individual fruiting bodies. Considering that juvenile specimens often had small immature cheilocystidia and pleurocystidia, the number of cystidia may be dependent on the age and the section of the fruiting body. Rickenella umbelliformis is phylogenetically closely related to R. indica. The two species differ in many morphological features, including the size of the pileus and pleurocystidia. Compared to the new species, Rickenella indica has a smaller pileus (1–7 mm) and longer pleurocystidia of 31–53 µm (Latha et al., 2015). The omphalinoid shape and the size of the fruiting body of R. umbelliformis are similar to those of a globally common Rickenella species, R. fibula (Bull.) Raithelh., but these species are well-divided by rDNA marker-based phylogenetic analyses. Micromorphological characteristics of R. fibula vary widely based on its geographical location, making it difficult to make a definite comparison.
Rigidoporaceae Jülich
Rigidoporus Murrill
Rigidoporus ginkgonis (Y.C. Dai) F. Wu, Jia J. Chen & Y.C. Dai, Fig. 5
MycoBank: MB 819205
Description: Basidiomes annual, resupinate, soft corky. Hymenophore porous, cream (4A4), fading to white to the margin; sterile margin indistinct, thinning out; pores round or angular, irregular, 4–5 per mm; dissepiments somewhat thin, entire; tubes up to 4.0 mm deep, concolorous with the pore surface.
Hyphal structure monomitic, generative hyphae thin-walled, hyaline. Basidiospores (4.0–)4.2–5.9 × 3.4–4.4(– 4.6) μm, L = 5.0 μm, W = 4.0 μm, Q = 1.1–1.4, globose to subglobose, thin-walled, hyaline, smooth, acyanophilous, and neither amyloid nor dextrinoid. Basidia 10.5–13.0(– 15.0) × 4.7–6.1 μm, broadly clavate to barrel-shaped, hyaline, 4-spored. Cystidia absent. Clamp connections absent.
Ecology/Substrate/Host: Grows on the trunk of living or dead Ginkgo biloba.
Distribution: China, Republic of Korea, and Uzbekistan
Specimen examined: Republic of Korea: Gangwon-do, Yanggu, Yanggu-eup, Godae-ri; 38°07′41.6″N, 127°59′24.7″E 181 m, sidewalk of Paro-ho, on the trunk of Ginkgo biloba; 30 June 2023, Young Woon Lim, Hannah Suh, Dohye Kim, Yoongil Lee (SFC20230630-23).
Notes: Morphological characters of Rigidoporus ginkgonis specimens from Korea match well with the morphological descriptions of the holotype. Only a few deviations exist, where the length of the tubes is shorter for the specimens from Korea (up to 4.5 mm for the holotype), and the size of the basidiospores is smaller compared to that of the holotype with (4.8–)5.0–6.0(– 6.5) × (3.9–)4.1–5.0(– 5.2) μm, L = 5.4 μm, W = 4.5 μm (Dai & Wang, 2005).
Sideraceae L.W. Zhou & Xue W. Wang.
Sidera Miettinen & K.H. Larss.
Sidera tibetica Z.B. Liu, Jian Yu & F. Wu, Fig. 6
MycoBank: MB 843516
Description: Basidiomes annual, resupinate, soft corky. Hymenophore porous, cream (4A3), fading to white to the margin; sterile margin indistinct, thinning out; pores round but somewhat angular, 8–9 per mm; dissepiments thin, entire to lacerate; tube concolorous with the pore surface.
Hyphal structure dimitic, skeletal hyphae dominant, thin-walled, hyaline, rarely branched, rosette-like crystals frequently present. Basidiospores (1.8–)2.0–2.6(– 2.8) × 0.5–0.9 μm, L = 2.3 μm, W = 0.7 μm, Q = 2.4–4.6, allantoid to cylindrical, thin-walled, hyaline, smooth, acyanophilous, and neither amyloid nor dextrinoid. Basidia (8.1–)8.6–9.6(– 10.1) × (3.5–)4.0–4.4 μm, barrel-shaped to clavate, hyaline, 4-spored. Cystidioles 9.4–12.7 × 2.6–3.9 μm, fusoid, basally swollen, somewhat sharp tip, infrequent, thin-walled, hyaline. Clamp connections present.
Ecology/Substrate/Host: Grows on dead Pinus spp.
Distribution: China, Republic of Korea
Specimen examined: Republic of Korea: Gwangju, Buk-gu, Mudeung-ro; 35°8′59″N 126°59′15″E, 360 m, Mt. Mudeung, mixed forest, on the trunk of a dead Pinus densiflora; 17 Mar 2023, Yoonhee Cho, Dohye Kim (SFC20230317-17).
Notes: Morphological characters of Sidera tibetica specimen from Korea largely correspond to those of the holotype. The only difference lies in the sizes of the basidiospores and cystidioles. The sizes were smaller for the specimen from Korea, where the holotype bore larger basidiospores of (2.8–)2.9–3.1(– 3.3) × 1–1.1(– 1.2) μm, and cystidioles of 13–15 × 3–4 μm (Liu et al., 2022).
Skvortzoviaceae L.W. Zhou & Xue W. Wang
Skvortzovia Bononi & Hjortstam
Skvortzovia dabieshanensis Jia Yu, Xue W. Wang, S.L. Liu & L.W. Zhou, Fig. 7
MycoBank: MB 840228
Description: Basidiomes annual, resupinate, closely adnate, not easily separable, thin, white to ash-gray (1B1). Hymenophore minutely grandinoid, 9–10 aculei per mm; margin thinning out, white.
Hyphal structure monomitic, hyaline, thin-walled, frequently branched. Basidiospores 4.5–4.6 × 1.7–1.9 μm, L = 4.6 μm, W = 1.8 μm, Q = 2.4–2.6, reniform, hyaline, smooth, acyanophilous, and neither amyloid nor dextrinoid. Basidia 10.0–14.9(– 20.9) × 3.1–4.2 μm, narrowly obpyriform, hyaline, 4-spored. Leptocystidia 17.8 × 3.5 μm, cylindrical, capitate, hyaline, thin-walled. Clamp connections present.
Ecology/Substrate/Host: Grows mostly on dead Pinus spp.
Distribution: China, Republic of Korea
Specimens examined: Republic of Korea: Gangwon-do, Yangyang-gun, Osaek-ri; 38°4′20″N 128°26′54″E, 415 m, Mt. Jeombong, Pinus koraiensis forest, on the trunk of a dead P. koraiensis; 7 May 2016, Young Woon Lim, Jae Young Park (SFC20160907-26). Republic of Korea: Gyeongsangnam-do, Hapcheon-gun, Mt. Gaya, Pinus densiflora forest, on the trunk of a dead P. densiflora; 9 Feb 2017, Young Woon Lim, Nam Kyu Kim, Hae Jin Cho (SFC20170209-12). Republic of Korea: Gyeongsangnam-do, Hapcheon-gun, Mt. Gaya, Pinus densiflora forest, on the trunk of a dead P. densiflora; 7 Sep 2018, Hyun Lee, Nam Hwi Kim, Abel Severin Lupala (SFC20180907-152). Republic of Korea: Gangwon-do, Inje-gun, Jindong-ri, on the trunk of a dead P. densiflora; 29 Sep 2018, Young Woon Lim, Ki Hyeong Park, Abel Severin Lupala (SFC20180929-32). Republic of Korea: Gyeongsangbuk-do, Bonghwa-gun, Seokpo-myeon, Daehyeon-ri; 37°4′16″N 128°57′60″E, Mt. Taebaek National Park, mixed forest, on the trunk of a dead P. densiflora; 22 Mar 2019, Young Woon Lim, Myung Soo Park, Min-Ji Kim, Hyun Lee, Ki Hyeong Park, Shinnam Yoo, Nam Hwi Kim (SFC20190322-05).
Notes: Most specimens of Skvortzovia dabieshanensis in Yu et al. (2021) and all our observed specimens developed basidiomes on Pinus as a host plant. Despite the geographical separation, specimens from Korea share similar morphological characteristics with those of the holotype. Still, they are distinctive in having smaller and more frequent aculei than the holotype, which bears 5–6 aculei per mm (Yu et al., 2021). Specimens from Korea also have narrower basidiospores and broader but shorter basidia than those of the holotype with basidiospores of (3.5–)3.8–4.7(− 4.9) × 1.8–2.4(− 2.6) μm and basidia of 10–20 × 3.5–5 μm (Yu et al., 2021).
Tubulicrinaceae Jülich
Tubulicrinis Donk
Tubulicrinis calothrix (Pat.) Donk, Fig. 8
MycoBank: MB 307182
Description: Basidiomes annual, resupinate, adnate, thin, discontinuous, rimulose, soft. Hymenophore smooth, white to light brown (4A3); margin indistinct, white, thinning out.
Hyphal structure monomitic, hyphae thin- to thick-walled, hyaline, rarely branched in subiculum, highly branched and septate in trama. Basidiospores (3.3–)3.7–4.9(– 5.0) × 1.1–1.9 μm, L = 4.3 μm, W = 1.5 μm, Q = 2.3–3.7, allantoid, thin-walled, hyaline, smooth, acyanophilous, and neither amyloid nor dextrinoid. Basidia (9.6–)9.7–12.3(– 13.5) × (2.7–)2.9–3.6(– 3.8) μm, subclavate, hyaline, 4-spored. Cystidia 50.0–88.1(– 93.4) × 4.8–9.9 μm, cylindrical to setiform, numerous, hyaline, some with bent and elongated base, thick-walled but narrowing to the obtuse and sometimes encrusted apex, capillary lumen abruptly expanded near the apex, amyloid. Clamp connections present.
Ecology/Substrate/Host: Commonly found in the northern and highland; grows on coniferous wood
Distribution: Widely reported across all continents except for South America
Specimen examined: Republic of Korea: Seoul, Gwanak-gu, Mt. Gwanak, mixed forest, on the trunk of a dead Pinus densiflora; 22 Aug 2018, Young Woon Lim, Abel Severin Lupala (SFC20180822-18).
Notes: There were very few fertile cells and basidiospores in tissue where cystidia were abundant, and vice versa. The specimen from Korea largely corresponded to the morphological characters described for the specimens from Northern Europe, but several micromorphological characters were disparate. Tubulicrinis calothrix in Korea has shorter basidiospores, smaller basidia, and shorter cystidia than those of T. calothrix from Northern Europe with basidiospores of 6–7(– 8) × 1.5–1.8(– 2) μm, basidia of 12–15 × 4–5 μm, and cystidia of 80–120 × 6–8 μm (Hjortstam et al., 1988).
Discussion
Species of Hymenochaetales families, apart from Hymenochaetaceae and Schizoporaceae, pose difficulties to study as they are often small, uncommon, or locality-specific (Nakasone & Burdsall, 2012), with some being endangered or threatened (Denchev et al., 2007; Karadelev & Rusevska, 2016; Tănase & Pop, 2005). Most of the understudied families were formerly placed under Rickenellaceae. Recently, it has been divided into several monogeneric families, i.e., Odonticiaceae, Peniophorellaceae, Resiniciaceae, Schizocorticiaceae, Sideraceae, and Skvortzoviaceae, based on multigenetic marker phylogeny and time divergence analysis (Wang et al., 2023).
The four resupinate unreported species, Rigidoporus ginkgonis, Sidera tibetica, Skvortzovia dabieshanensis, and Tubulicrinis calothrix may be easily mistaken as other species in the field as their insubstantial macromorphological characters overlap with other corticoid or polyporous species of Hymenochaetales and other families. In fact, before molecular work was introduced in fungal taxonomy, Tubulicrinis calothrix was reported as Corticium based on its effused basidiome, cream hymenophore, powdery margin, and abundant large cystidia (Patouillard et al., 1897). After its transfer to Tubulicrinis by Donk (1956), Tubulicrinis was only later verified through nLSU for its placement in Hymenochaetales (Larsson et al., 2006). Similarly, Rigidoporus ginkgonis, like many other Rigidoporus species, has been initially reported under Oxyporus based on morphology but has undergone a systematic transition later based on phylogeny (Wu et al., 2017). Therefore, it is incumbent for researchers to validate the classification and identification of many species with poor morphological traits through phylogenetic analyses.
Small bryophilous orange omphaloid mushrooms have been repeatedly reported as Rickenella fibula in Korea based on their macromorphology (Jo et al., 2013; Kim et al., 2017). However, our phylogenetic analysis presents three Rickenella species in the country — R. fibula, R. indica, and R. umbelliformis sp. nov. (Figs. 1 and 2, Fig. S2). As Rickenella species are challenging to identify based on morphology due to overlapping characters, phylogenetic support is essential. Rickenella species possess significant values for ecological and evolutionary research as they consist of a wide range of morphological characters and lifestyles, from saprotrophic to bryicolous, that may be symbiotic or parasitic to the hosts (Bresinsky & Schötz, 2006; Korotkin et al., 2018). However, prior to other research, a concrete taxonomic study is inevitable for the genus as some Rickenella species are not phylogenetically well resolved. The clades of R. setipes and R. danxiashanensis are nested within R. swartzii and R. mellea, respectively. Rickenella fibula is paraphyletic, and two new species candidates were detected, annotated as Rickenella sp. 1 and 2, respectively (Fig. 2).
Despite how infallible phylogenetic analysis may seem, morphological assessment is essential when differentiating phylogenetically closely related species. Peniophorella specimens analyzed in the present study were grouped monophyletically with both P. odontiiformis and P. rude (Fig. 1). However, they were eventually identified as P. odontiiformis based on basidiospore measurements (Table S1). The basidiospore measurements of the studied specimens closely matched the range of P. odontiiformis (7.5–9 × 3.5–5 μm) than that of P. rude (9–10 × 6 μm) (Larsson, 2007). This problem arose partially due to some inaccurate GenBank annotations, such as DQ647494 and DQ647497, that were annotated as P. rude when they were identified as P. odontiiformis in the reference research article (Hallenberg et al., 2007). This stipulates the need to evaluate morphological characters in addition to molecular analysis for accurate species identification.
A consistent effort to update indigenous species to the most recent taxonomic classification is imperative. This is because various problems may arise from the persistent use of old names or synonyms, such as losing track of type specimens, misidentification, and inconsistent ecological and morphological descriptions of species (Dayarathne et al., 2016). Trichaptum has long been classified as incertae sedis (uncertain placement) due to its undecisive taxonomic position at the family level within Hymenochaetales. Recently, Zhou et al. (2023) resurrected Trichaptum sensu stricto to Trichaptaceae Y.C. Dai, Yuan Yuan & Meng Zhou, and introduced another new family, Hirschioporaceae Y.C. Dai, Yuan Yuan & Meng Zhou, to encompass some species in Trichaptum sensu lato. Based on our results, all Trichaptum specimens in Korea are now classified under Hirschioporaceae as Hirschioporus or Pallidohirschioporus species (Fig. 1). As such, there is currently no Trichaptum species in the country until a new discovery occurs. As Trichaptum sensu lato species are documented and subjected to comprehensive research across diverse research areas in Korea (Cho et al., 2020; Kim et al., 1998; Lupala et al., 2019), it is critical to contemplate taxonomic revisions to prevent potential confusion across all studies.
In conclusion, the multistage morphological and multigenetic marker-based phylogenetic investigation revealed 18 species in eight understudied Hymenochaetales families (exclusive of Hymenochaetaceae and Schizoporaceae) from Korea. The most recent taxonomic classification and revisions were reflected upon these species. The national species list will extend with continuous findings and research. Limited specimen records and genetic marker sequences are available for the species under incertae sedis. As most understudied species possess inconspicuous fruiting bodies, a more attentive fruiting body collection and genetic marker sequence production are required to expand and enrich the taxonomic studies of Hymenochaetales. The effort will further fill the gaps of unsupported taxa and eventually assign all genera and species to the appropriate classification.
Data availability
The study materials and data from this study are available from the corresponding author upon reasonable request.
References
Bresinsky, A., & Schötz, A. (2006). Behaviour in cultures and habitat requirements of species within the genera Loreleia and Rickenella (Agaricales). Acta Mycologica, 41, 189–208.
Cho, S. E., Kwag, Y. N., Jo, J. W., Han, S. K., Oh, S. H., & Kim, C. S. (2020). Macrofungal diversity of urbanized areas in southern part of Korea. Journal of Asia-Pacific Biodiversity, 13, 189–197.
Cho, Y., Kim, D., Lee, Y., Jeong, J., Hussain, S., & Lim, Y. W. (2023). Validation of Fuscoporia (Hymenochaetales, Basidiomycota) ITS sequences and five new species based on multi-marker phylogenetic and morphological analyses. IMA Fungus, 14, 12.
Cho, Y., Kim, J. S., Dai, Y. C., Gafforov, Y., & Lim, Y. W. (2021). Taxonomic evaluation of Xylodon (Hymenochaetales, Basidiomycota) in Korea and sequence verification of the corresponding species in GenBank. PeerJ, 9, e12625.
Dai, Y. C., & Wang, Z. (2005). A new polypore on Ginkgo from China. Mycotaxon, 92, 345–349.
Dayarathne, M. C., Boonmee, S., Braun, U., Crous, P. W., Daranagama, D. A., Dissanayake, A. J., Ekanayaka, H., Jayawardena, R., Jones, E. B. G., Maharachchikumbura, S. S. N., et al. (2016). Taxonomic utility of old names in current fungal classification and nomenclature: Conflicts, confusion & clarifications. Mycosphere, 7, 1622–1648.
Denchev, C. M., Fakirova, V. I., Gyosheva, M. M., & Petrova, R. D. (2007). Macromycetes in the Pirin Mts (SW Bulgaria). Acta Mycologica, 42, 21–34.
Dentinger, B. T., & McLaughlin, D. J. (2006). Reconstructing the Clavariaceae using nuclear large subunit rDNA sequences and a new genus segregated from Clavaria. Mycologia, 98, 746–762.
Donk, M. A. (1956). Notes on resupinate Hymenomycetes–III. Fungus, 26, 3–24.
Gardes, M., & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology, 2, 113–118.
Hallenberg, N., Nilsson, R. H., Antonelli, A., Wu, S. H., Maekawa, N., & Nordén, B. (2007). The Peniophorella praetermissa species complex (Basidiomycota). Mycological Research, 111, 1366–1376.
Hibbett, D. S., & Thorn, R. G. (2001). The fungal hierarchy: 5. Homobasidiomycetes. In D. J. McLaughlin, E. G. McLaughlin, & P. A. Lemke (Eds.), Systematics and Evolution. The Mycota. (Vol. 7B). Springer.
Hjortstam, K., Larsson, K. H., Ryvarden, L., & Eriksson, J. (1988). The Corticiaceae of North Europe (Vol. 8, pp. 1450–1631). Fungiflora.
Huelsenbeck, J. P., & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Jang, Y., Jang, S., Lee, J., Lee, H., Lim, Y. W., Kim, C., & Kim, J. J. (2016). Diversity of wood-inhabiting polyporoid and corticioid fungi in Odaesan National Park, Korea. Mycobiology, 44, 217–236.
Jo, J. W., Kwag, Y. N., Kim, C. S., & Han, S. K. (2013). Distribution of higher fungi in Chungcheong province, Republic of Korea. Journal of Asia-Pacific Biodiversity, 6, 347–359.
Karadelev, M., & Rusevska, K. (2016). Distribution maps of critical endangered species from Macedonian Red List of Fungi. Hyla, 2016, 14–18.
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780.
Kim, B. K., Choi, E. C., Chung, K. S., & Lee, Y. S. (1976). Taxonomic investigations on Korean higher fungi (III). Korean Journal of Pharmacognosy, 7, 199–208.
Kim, C. Y., Jang, S. K., & Kim, M. S. (2017). Occurrence according to resource utilization characteristics of higher fungi in Naejangsan National Park. Korean Journal of Mycology, 45, 270–283.
Kim, C., Lee, J. S., Jung, H. S., & Lim, Y. W. (2009). The wood-rotting fungal flora of three islands in the Yellow Sea, Korea. Mycobiology, 37, 147–151.
Kim, J. H., Lee, H. Y., Yoo, K. H., Kim, Y. S., Seok, S. J., & Kim, Y. S. (1998). The screening of fibrinolytic activities of extracts from mushrooms in Mt. Chiak. The Korean Journal of Mycology, 26, 589–593.
Kim, N. K., Park, J. Y., Park, M. S., Lee, H., Cho, H. J., Eimes, J. A., Kim, C., & Lim, Y. W. (2016). Five new wood decay fungi (Polyporales and Hymenochaetales) in Korea. Mycobiology, 44, 146–154.
Kirk, P. M., Cannon, P. F., Minter, D. W., & Stalpers, J. A. (2008). Dictionary of the Fungi (10th ed.). CAB International.
Kornerup, A., & Wanscher, J. (1978). Methuen handbook of colour (3rd ed.). Eyre Methuen.
Korotkin, H. B., Swenie, R. A., Miettinen, O., Budke, J. M., Chen, K. H., Lutzoni, F., Smith, M. E., & Matheny, P. B. (2018). Stable isotope analyses reveal previously unknown trophic mode diversity in the Hymenochaetales. American Journal of Botany, 105, 1869–1887.
Larsson, K. H. (2007). Molecular phylogeny of Hyphoderma and the reinstatement of Peniophorella. Mycological Research, 111, 186–195.
Larsson, K. H., Parmasto, E., Fischer, M., Langer, E., Nakasone, K. K., & Redhead, S. A. (2006). Hymenochaetales: A molecular phylogeny for the hymenochaetoid clade. Mycologia, 98, 926–936.
Latha, K. P. D., Raj, K. N. A., Paramban, R., & Manimohan, P. (2015). Two new bryophilous agarics from India. Mycoscience, 56, 75–80.
Liu, Z. B., Zhou, M., Wu, F., & Yu, J. (2022). Two new species of Sidera (Hymenochaetales, Basidiomycota) from Southwest China. Journal of Fungi, 8, 385.
Liu, Z. B., Zhou, M., Yuan, Y., & Dai, Y. C. (2021). Global diversity and taxonomy of Sidera (Hymenochaetales, Basidiomycota): Four new species and keys to species of the genus. Journal of Fungi, 7, 251.
Lupala, A. S., Oh, S. Y., Park, M. S., Kim, T., Yoo, J. S., Seelan, J. S. S., & Lim, Y. W. (2019). Co-occurrence patterns of wood-decaying fungi and ants in dead pines of South Korea. Journal of Asia-Pacific Entomology, 22, 1154–1160.
Matheny, P. B. (2005). Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Molecular Phylogenetics and Evolution, 35, 1–20.
Nakasone, K., & Burdsall, H. (2012). Tsugacorticium kenaicum (Hymenochaetales, Basidiomycota), a new corticioid genus and species from Alaska. North Am Fungi, 7, 1–9.
Park, M. S., Cho, H. J., Kim, N. K., Park, J. Y., Lee, H., Park, K. H., Kim, M. J., Kim, J. J., Kim, C., & Lim, Y. W. (2017). Ten new recorded species of Macrofungi on Ulleung Island, Korea. Mycobiology, 45, 286–296.
Patouillard, N., Barratte, G., & Sauvageau, C. (1897). Catalogue raisonné des plantes cellulaires de la Tunisie. Paris, France: Imprimerie Nationale.
Redhead, S. A., Moncalvo, J. M., Vilgalys, R., & Lutzoni, F. (2002). Phylogeny of agarics: Partial systematics solutions for bryophilous omphalinoid agarics outside of the Agaricales (Euagarics). Mycotaxon, 82, 151–168.
Rehner, S. A., & Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia, 97, 84–98.
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313.
Tănase, C., & Pop, A. (2005). Red list of Romanian macrofungi species. In S. Mihãilescu (Ed.), Bioplatform-romanian national platform for biodiversity (pp. 101–107). Editura Academiei Române.
Tedersoo, L., Suvi, T., Beaver, K., & Saar, I. (2007). Ectomycorrhizas of Coltricia and Coltriciella (Hymenochaetales, Basidiomycota) on Caesalpiniaceae, Dipterocarpaceae and Myrtaceae in Seychelles. Mycological Progress, 6, 101–107.
van Driel, K. G., Humbel, B. M., Verkleij, A. J., Stalpers, J., Müller, W. H., & Boekhout, T. (2009). Septal pore complex morphology in the Agaricomycotina (Basidiomycota) with emphasis on the Cantharellales and Hymenochaetales. Mycological Research, 113, 559–576.
Vilgalys, R., & Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology, 172, 4239–4246.
Wang, X. W., Liu, S. L., & Zhou, L. W. (2023). An updated taxonomic framework of Hymenochaetales (Agaricomycetes, Basidiomycota). Mycosphere, 14, 452–496.
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplifications and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfland, J. Sninsky, & T. J. White (Eds.), PCR Protocols: A guide to methods and applications (pp. 315–322). Academic Press.
Wu, F., Chen, J. J., Ji, X. H., Vlasák, J., & Dai, Y. C. (2017). Phylogeny and diversity of the morphologically similar polypore genera Rigidoporus, Physisporinus, Oxyporus, and Leucophellinus. Mycologia, 109, 749–765.
Yu, J., Wang, X. W., Liu, S. L., Shen, S., & Zhou, L. W. (2021). Taxonomy and phylogeny of Resinicium sensu lato from Asia-Pacific revealing a new genus and five new species (Hymenochaetales, Basidiomycota). IMA Fungus, 12, 19.
Zhang, M., Li, T. H., & Chen, F. (2018). Rickenella danxiashanensis, a new bryophilous agaric from China. Phytotaxa, 350, 174–175.
Zhou, M., Dai, Y. C., Vlasák, J., Liu, H. G., & Yuan, Y. (2023). Revision and updated systematics of Trichaptum s.l. (Hymenochaetales, Basidiomycota). Mycosphere, 14, 815–917.
Acknowledgements
This study was supported by the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of Korea (NIBR202304104).We express our deep gratitude to Dr. Seung Se Choi (National Institute of Ecology, KR) for his assistance in identifying bryophytes associated with Rickenella species.
Funding
Open Access funding enabled and organized by Seoul National University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
12275_2024_120_MOESM1_ESM.fasta
Supplementary Data 1. Concatenated nSSU+ ITS+ nLSU+ RPB2+ TEF1 dataset for multigenetic marker phylogenetic analysis of Hymenochaetales
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cho, Y., Kim, D. & Lim, Y.W. Phylogenetic Assessment of Understudied Families in Hymenochaetales (Basidiomycota, Fungi)—Reporting Uncovered Species and Reflecting the Recent Taxonomic Updates in the Republic of Korea. J Microbiol. 62, 429–447 (2024). https://doi.org/10.1007/s12275-024-00120-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-024-00120-5