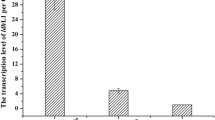
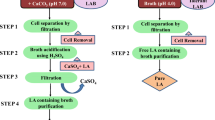
Abstract
D-Lactic acid is a chiral, three-carbon organic acid, that bolsters the thermostability of polylactic acid. In this study, we developed a microbial production platform for the high-titer production of D-lactic acid. We screened 600 isolates of lactic acid bacteria (LAB) and identified twelve strains that exclusively produced D-lactic acid in high titers. Of these strains, Lactobacillus saerimneri TBRC 5746 was selected for further development because of its homofermentative metabolism. We investigated the effects of high temperature and the use of cheap, renewable carbon sources on lactic acid production and observed a titer of 99.4 g/L and a yield of 0.90 g/g glucose (90% of the theoretical yield). However, we also observed L-lactic acid production, which reduced the product’s optical purity. We then used CRISPR/dCas9-assisted transcriptional repression to repress the two Lldh genes in the genome of L. saerimneri TBRC 5746, resulting in a 38% increase in D-lactic acid production and an improvement in optical purity. This is the first demonstration of CRISPR/dCas9-assisted transcriptional repression in this microbial host and represents progress toward efficient microbial production of D-lactic acid.







Similar content being viewed by others
References
Abdel-Rahman, M. A., & Sonomoto, K. (2016). Opportunities to overcome the current limitations and challenges for efficient microbial production of optically pure lactic acid. Journal of Biotechnology, 236, 176–192.
Abdel-Rahman, M. A., Tashiro, Y., & Sonomoto, K. (2013). Recent advances in lactic acid production by microbial fermentation processes. Biotechnology Advances, 31, 877–902.
Aguirre-Ezkauriatza, E. J., Aguilar-Yáñez, J. M., Ramírez-Medrano, A., & Alvarez, M. M. (2010). Production of probiotic biomass (Lactobacillus casei) in goat milk whey: Comparison of batch, continuous and fed-batch cultures. Bioresource Technology, 101, 2837–2844.
Alves de Oliveira, R., Komesu, A., Rossell, C. E. V., & Maciel Filho, R. (2018). Challenges and opportunities in lactic acid bioprocess design—From economic to production aspects. Biochemical Engineering Journal, 133, 219–239.
Bae, S., Park, J., & Kim, J. S. (2014). Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics, 30, 1473–1475.
Bai, D. M., Wei, Q., Yan, Z. H., Zhao, X. M., Li, X. G., & Xu, S. M. (2003). Fed-batch fermentation of Lactobacillus lactis for hyper-production of L-lactic acid. Biotechnology Letters, 25, 1833–1835.
Bai, Z., Gao, Z., Sun, J., Wu, B., & He, B. (2016). D-Lactic acid production by Sporolactobacillus inulinus YBS1-5 with simultaneous utilization of cottonseed meal and corncob residue. Bioresource Technology, 207, 346–352.
Bai, H., Deng, S., Bai, D., Zhang, Q., & Fu, Q. (2017). Recent Advances in Processing of Stereocomplex-Type Polylactide. Macromolecular Rapid Communications, 38, 1700454.
Castro-Aguirre, E., Iniguez-Franco, F., Samsudin, H., Fang, X., & Auras, R. (2016). Poly(lactic acid)—Mass production, processing, industrial applications, and end of life. Advanced Drug Delivery Reviews, 107, 333–366.
Chen, N., Wang, J., Zhao, Y., & Deng, Y. (2018). Metabolic engineering of Saccharomyces cerevisiae for efficient production of glucaric acid at high titer. Microbial Cell Factories, 17, 67.
Choi, Y. J., & Lee, S. Y. (2013). Microbial production of short-chain alkanes. Nature, 502, 571–574.
Coelho, L. F., de Lima, C. J., Bernardo, M. P., & Contiero, J. (2011). D(-)-lactic acid production by Leuconostoc mesenteroides B512 using different carbon and nitrogen sources. Applied Biochemistry and Biotechnology, 164, 1160–1171.
Dominguez, A. A., Lim, W. A., & Qi, L. S. (2016). Beyond editing: repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nature Reviews Molecular Cell Biology, 17, 5–15.
Doudna, J. A., & Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science, 346, 1258096.
Gänzle, M. G., & Follador, R. (2012). Metabolism of oligosaccharides and starch in lactobacilli: a review. Frontiers in Microbiology, 3, 340.
Gu, S. A., Jun, C., Joo, J. C., Kim, S., Lee, S. H., & Kim, Y. H. (2014). Higher thermostability of L-lactate dehydrogenases is a key factor in decreasing the optical purity of D-lactic acid produced from Lactobacillus coryniformis. Enzyme and Microbial Technology, 58–59, 29–35.
Huang, H., Song, X., & Yang, S. (2019). Development of a RecE/T-Assisted CRISPR–Cas9 Toolbox for Lactobacillus. Biotechnology Journal, 14, e1800690.
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821.
Jun, C., Sa, Y. S., Gu, S. A., Joo, J. C., Kim, S., Kim, K. J., & Kim, Y. H. (2013). Discovery and characterization of a thermostable D-lactate dehydrogenase from Lactobacillus jensenii through genome mining. Process Biochemistry, 48, 109–117.
Juturu, V., & Wu, J. C. (2016). Microbial production of lactic acid: The latest development. Critical Reviews in Biotechnology, 36, 967–977.
Kim, S., Gu, S. A., Kim, Y. H., & Kim, K. J. (2014). Crystal structure and thermodynamic properties of D-lactate dehydrogenase from Lactobacillus jensenii. International Journal of Biological Macromolecules, 68, 151–157.
Kruyer, N. S., & Peralta-Yahya, P. (2017). Metabolic engineering strategies to bio-adipic acid production. Current Opinion in Biotechnology, 45, 136–143.
Lahtinen, S., Ouwehand, A. C., Salminen, S., & von Wright, A. (2012). Lactic Acid Bacteria: Microbiological and Functional Aspects (4th ed.). CRC Press.
Leenay, R. T., Vento, J. M., Shah, M., Martino, M. E., Leulier, F., & Beisel, C. L. (2019). Genome editing with CRISPR-Cas9 in Lactobacillus plantarum revealed that editing outcomes can vary across strains and between methods. Biotechnology Journal, 14, e1700583.
Liao, J. C., Mi, L., Pontrelli, S., & Luo, S. (2016). Fuelling the future: microbial engineering for the production of sustainable biofuels. Nature Reviews Microbiology, 14, 288–304.
Loubiere, P., Cocaign-Bousquet, M., Matos, J., Goma, G., & Lindley, N. D. (1997). Influence of end-products inhibition and nutrient limitations on the growth of Lactococcus lactis subsp. lactis. Journal of Applied Microbiology, 82, 95–100.
Luedeking, R., & Piret, E. L. (1959). A kinetic study of the lactic acid fermentation. Batch process at controlled pH. Biotechnology and Bioengineering, 1, 393–412.
Moon, T. S., Yoon, S. H., Lanza, A. M., Roy-Mayhew, J. D., & Prather, K. L. (2009). Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Applied and Environmental Microbiology, 75, 589–595.
Othman, M., Ariff, A. B., Rios-Solis, L., & Halim, M. (2017). Extractive fermentation of lactic acid in lactic acid bacteria cultivation: a review. Frontiers in Microbiology, 8, 2285.
Park, J., Bae, S., & Kim, J. S. (2015). Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics, 31, 4014–4016.
Reid, S. J., & Abratt, V. R. (2005). Sucrose utilisation in bacteria: genetic organisation and regulation. Applied Microbiology and Biotechnology, 67, 312–321.
Romano, A., Trip, H., Campbell-Sills, H., Bouchez, O., Sherman, D., Lolkema, J. S., & Lucas, P. M. (2013). Genome sequence of Lactobacillus saerimneri 30a (Formerly Lactobacillus sp. Strain 30a), a reference lactic acid bacterium strain producing biogenic amines. Genome Announcements, 1, e00097-e112.
Sornlek, W., Sae-Tang, K., Watcharawipas, A., Wongwisansri, S., Tanapongpipat, S., Eurwilaichtr, L., Champreda, V., Runguphan, W., Schaap, P. J., & Martins Dos Santos, V. A. P. (2022). D-Lactic acid production from sugarcane bagasse by genetically engineered Saccharomyces cerevisiae. Journal of Fungi, 8, 816.
Watcharawipas, A., Sae-Tang, K., Sansatchanon, K., Sudying, P., Boonchoo, K., Tanapongpipat, S., Kocharin, K., & Runguphan, W. (2021). Systematic engineering of Saccharomyces cerevisiae for D-lactic acid production with near theoretical yield. FEMS Yeast Research, 21, foab024.
Wu, J., Xin, Y., Kong, J., & Guo, T. (2021). Genetic tools for the development of recombinant lactic acid bacteria. Microbial Cell Factories, 20, 118.
Acknowledgements
The project was funded by the Integrated Technology Platform (Biobased Materials) project, “IBMDL1-High-level Microbial Production of Enantiomerically Pure D-lactic Acid,” supported by the National Science and Technology Development Agency [grant number P-17-522777].
Author information
Authors and Affiliations
Contributions
Conceptualization, WR, WV and KK; methodology, WR and KK; validation, KS, PS, YK, KK and WR; formal analysis, KK and WR; investigation, KS, PS and PP; resources, WR; data curation, KS, PS, YK, KK and WR; writing—original draft preparation, WR and KK; writing—review and editing, WR, KK and PP; visualization, WR and KK; supervision, WV, ST, WR and KK; project administration, WR and KK; funding acquisition, WR All authors have read and agreed to the published version of the manuscript.”
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest. The funder had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Ethical approval
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sansatchanon, K., Sudying, P., Promdonkoy, P. et al. Development of a Novel D-Lactic Acid Production Platform Based on Lactobacillus saerimneri TBRC 5746. J Microbiol. 61, 853–863 (2023). https://doi.org/10.1007/s12275-023-00077-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-023-00077-x