Abstract
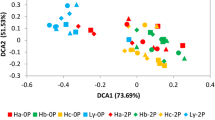
Microbial communities in the rhizosphere play a crucial role in determining plant growth and crop yield. A few studies have been performed to evaluate the diversity and co-occurrence patterns of rhizosphere microbiomes in soybean (Glycine max) at a regional scale. Here, we used a culture-independent method to compare the bacterial communities of the soybean rhizosphere between Nebraska (NE), a high-yield state, and Oklahoma (OK), a low-yield state. It is well known that the rhizosphere microbiome is a subset of microbes that ultimately get colonized by microbial communities from the surrounding bulk soil. Therefore, we hypothesized that differences in the soybean yield are attributed to the variations in the rhizosphere microbes at taxonomic, functional, and community levels. In addition, soil physicochemical properties were also evaluated from each sampling site for comparative study. Our result showed that distinct clusters were formed between NE and OK in terms of their soil physicochemical property. Among 3 primary nutrients (i.e., nitrogen, phosphorus, and potassium), potassium is more positively correlated with the high-yield state NE samples. We also attempted to identify keystone communities that significantly affected the soybean yield using co-occurrence network patterns. Network analysis revealed that communities formed distinct clusters in which members of modules having significantly positive correlations with the soybean yield were more abundant in NE than OK. In addition, we identified the most influential bacteria for the soybean yield in the identified modules. For instance, included are class Anaerolineae, family Micromonosporaceae, genus Plantomyces, and genus Nitrospira in the most complex module (ME9) and genus Rhizobium in ME23. This research would help to further identify a way to increase soybean yield in low-yield states in the U.S. as well as worldwide by reconstructing the microbial communities in the rhizosphere.
Similar content being viewed by others
Data Availability
The complete 16S rRNA gene sequencing data in this study are available in the National Center for Biotechnology Information (NCBI) database under the BioProject accession number PRJNA873129 associated with the accession numbers of 12 BioSamples (SAMN30489248 - SAMN30489259).
References
Barberán, A., Bates, S.T., Casamayor, E.O., and Fierer, N. 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J., 6, 343–351.
Bastian, M., Heymann, S., and Jacomy, M. 2009. Gephi: an open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media, 3, 361–362. Retrieved from https://ojs.aaai.org/index.php/ICWSM/article/view/13937.
Becerra-Rivera, V.A. and Dunn, M.F. 2019. Polyamine biosynthesis and biological roles in rhizobia. FEMS Microbiol. Rev., 366, fnz084.
Berry, D. and Widder, S. 2014. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol., 5, 219.
Bokulich, N.A., Kaehler, B.D., Rideout, J.R., Dillon, M., Bolyen, E., Knight, R., Huttley, G.A., and Caporaso, J.G. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome, 6, 90.
Bolyen, E., Rideout, J.M., Dillon, M.R., Bokulich, N.A., Abnet, C.C., Al Ghalith, A.G., Alexander, H., Alm, E.J., Arumugam, M., Asnicar, F., et al. 2018. QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. Nat. Biotechnol., 37, 852–857.
Boukhatem, Z.F., Merabet, C., and Tsaki, H. 2022. Plant growht promoting actinobacteria, the most promising candidates as bioinoculants?. Front. Agron., 4, 849911.
Cao Y. 2022. Package ‘microbiomeMarker’: microbiome biomarker analysis toolkit. R package version 1.2.2. https://github.com/yiluheihei/microbiomeMarker.
Chi, S.C., Mothersole, D.J., Dilbeck, P., Niedzwiedzki, D.M., Zhang, H., Qian, P., Vasilev, C., Grayson, K.J., Jackson, P.J., Martin, E.C., et al. 2015. Assembly of functional photosystem complexes in Rhodobacter sphaeroides incorporating carotenoids from the spirilloxanthin pathway. Biochim. Biophys. Acta, 1847, 189–201.
Chowdhury, C., Sinha, S., Chun, S., Yeates, T.O., and Bobik, T.A. 2014. Diverse bacterial microcompartment organelles. Microbiol. Mol. Biol. Rev., 78, 438–468.
Clarke, K.R. and Ainsworth, M. 1993. A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser., 92, 205–219.
Collins, M.D. and Jones, D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol. Rev., 45, 316–354.
Csárdi, G. and Nepusz, T. 2006. The igraph software package for complex network research. InterJ. Complex Syst., 1695, 1–9.
Cram, J.A., Xia L.C., Needham D.M., Sachdeva R., Sun F., and Fuhrman J.A. 2015. Cross-depth analysis of marine bacterial networks suggests downward propagation of temporal changes. ISME J., 9, 2573–2586.
de Menezes, A.B., Prendergast-Miller, M.T., Richardson, A.E., Toscas, P., Farrell, M., Macdonald, L.M., Baker, G., Wark, T., and Thrall, P.H. 2015. Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ. Microbiol., 17, 2677–2689.
Ding, B., Niu, J., Shang, F., Yang, L., Chang, T., and Wang, J. 2019. Characterization of the geranylgeranyl diphosphate synthase gene in Acyrthosiphon pisum (Hemiptera: Aphididae) and its association with carotenoid biosynthesis. Front. Physiol., 10, 1398.
Douglas, G.M., Maffei, V.J., Zaneveld, J., Yurgel, S.N., Brown, J.R., Taylor, C.M., Huttenhower, C., Langille, M.G.I. 2020. PICRUSt2: an improved and extensible approach for metagenome inference. Nat. Biotechnol., 38, 685–688.
El-Tarabily, K., AlKhajeh, A.S., Ayyash, M.M., Alnuaimi, L.H., Sham, A., ElBaghdady, K.Z., Tariq, S., and AbuQamar, S.F. 2019. Growth promotion of Salicornia bigelovii by Micromonospora chalcea UAE1, and endophytic 1-aminocyclopropane-1-carboxylic acid deaminase-producing actinobacterial isolate. Front. Microbiol., 10, 1694.
Expósito, R.G., Postma, J., Raaijmakers, J.M., and Bruijn, I.D. 2015. Diversity and activity of Lysobacter species from disease suppressive soils. Front. Microbiol., 6, 1243.
Faust, K. and Raes, J. 2012. Microbial interactions: from networks to models. Nat. Rev. Microbiol., 10, 538–550.
Fierer, N. and Jackson, R.B. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA, 103, 626–631.
Hartmann, A., Rothballer, M., and Schmid, M. 2008. Lorenz Hiltner; a pioneer in rhizosphere microbial ecology & soil bacteriology research. Plant Soil, 312, 7–14.
Herridge, D.F., Peoples, M.B., and Boddey, R.M. 2008. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil, 311, 1–18.
Hobley, L., Kim, S.H., Maezato, Y., Wyllie, S., Fairlamb, A.H., Stanley-Wall, N.R., and Michael, A.J. 2014. Norspermidine is not a selfproduced trigger for biofilm disassembly. Cell, 156, 844–854.
Jiménez, J.A., Novinscak, A., and Filion, M. 2020. Inoculation with plant-growth-promoting rhizobacterium Pseudomonas fluorescens LBUM677 impacts the rhizosphere microbiome of three oilseed crops. Front. Microbiol., 11, 569366.
Jin, D., Kong, X., Li, H., Luo, L., Zhuang, X., Zhuang, G., Deng, Y., and Bai, Z. 2016. Patulibacter brassicae sp. nov., isolated from rhizosphere soil of chinese cabbage (Brassica campestris). Int. J. Syst. Evol. Microbiol., 66, 5056–5060.
Jin, J., Wang, G.H., Liu, X.B., Liu, J.D., Chen, X.L., and Herbert, S.J. 2009. Temporal and spatial dynamics of bacterial community in the rhizosphere of soybean genotypes grown in a black soil. Pedosphere, 19, 808–816.
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res., 30, 3059–3066.
Killiny, N. and Nehela, Y. 2020. Citrus polyamines: structure, biosynthesis, and physiological functions. Plants, 9, 426.
Kim, H.S., Kim, Y.H., Yoo, O.J., and Lee, J.J. 1996. Aclacinomycin X, a novel anthracycline antibiotic produced by Streptomyces galilaeus ATCC 31133. Biosci. Biotech. Biochem., 60, 906–908.
Kraut-Cohen, J., Shapiro, O.H., Dror, B., and Cytryn, E. 2021. Pectin induced colony expansion of soil-derived Flavobacterium strains. Front. Microbiol., 12, 651891.
Kuffner, M., Puschenreiter, M., Wieshammer, G., Gorfer, M., and Sessitsch, A. 2008. Rhizosphere bacteria affect growth and metal uptake of heavy metal accumulating willows. Plant Soil, 304, 35–44.
Langfelder, P. and Horvath, S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559.
Langille, M.G.I., Zaneveld, J., Caporaso, J.G., McDonald, D., Knights, D., Reyes, J.A., Clemente, J.C., Burkepile, D.E., Thurber, R.L.V., Knight, R., et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol., 31, 814–821.
Lankau, R.A., George, I., and Miao, M. 2022. Crop performance is predicted by soil microbial diversity across phylogenetic scales. Ecosphere, 13, e4029.
Liang, J., Sun, S., Ji, J., Wu, H., Meng, F., Zhang, M., Zheng, X., Wu, C., and Zhang, Z. 2014. Comparison of the rhizosphere bacterial communities of zigongdongdou soybean and a high-methionine transgenic line of this cultivar. PLoS ONE, 9, e103343.
Liu, C., Sun, Z., Shen, S., Lin, L., Li, T., Tian, B., and Hua, Y. 2013. Identification and characterization of the geranylgeranyl diphosphate synthase in Deinococcus radiodurans. Lett. Appl. Microbiol., 58, 219–224.
Lozupone, C.A., Hamady, M., Kelley, S.T., and Knight, R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol., 73, 1576–1585.
Lozupone, C., Lladser, M., Knights, D., Stombaugh, J., and Knight, R. 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J., 5, 169–172.
Luster, J., Göttlein, A., Nowack, B., and Sarret, G. 2009. Sampling, defining, characterising and modeling the rhizosphere-the soil science tool box. Plant Soil, 321, 457–482.
Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res., 27, 209–220.
Mcdonald, D., Price, M.N., Goodrich, J., Nawrocki, E.P., DeSantis, T.Z., Probst, A., Andersen, G.L., Knight, R., and Hugenholtz, P. 2012. An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME, J. 6, 610–618.
Mehrani, M., Sobotka, D., Kowal, P., Ciesielski, S., and Makinia, J. 2020. The occurrence and role of Nitrospiria in nitrogen removal systems. Bioresour. Technol., 303, 122936.
Mendes, L.W., Kuramae, E.E., Navarrete, A.A., van Veen, J.A., and Tsai, S.M. 2014. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J., 8, 1577–1587.
Meng, J., Xu, Y., Li, S., Li, C., Zhang, X., Dong, D., and Chen, P. 2010. Soybean growth and soil microbial populations under conventional and conservational tillage systems. J. Crop Improv., 24, 337–348.
Mizrahi-Man, O., Davenport, E.R., and Gilad, Y. 2013. Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS ONE, 8, e53608.
Naik, D., Smith, E., and Cumming, J.R. 2009. Rhizosphere carbon deposition, oxidative stress and nutritional changes in two poplar species exposed to aluminum. Tree Physiol., 29, 423–436.
Naim, M.S. 1965. Development of rhizosphere and rhizoplane microflora of Aristida coerulescens in the Libyan desert. Archiv. Mikrobiol., 50, 321–325.
Newman, M.E.J. and Girvan, M. 2004. Finding and evaluating community structure in networks. Phys. Rev. E 69, 026113.
Niraula, S., Choi, Y.K., Payne, K., Muir, J.P., Kan, E., and Chang, W.S. 2021. Dairy effluent-saturated biochar alters microbial communities and enhances bermudagrass growth and soil fertility. Agronomy, 11, 1794.
Nowicka, B. and Kruk, J. 2010. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta, 1797, 1587–1605.
Park, D., Kim, H., and Yoon, S. 2017. Nitrous oxide reduction by an obligate aerobic bacterium, Gemmatimonas aurantiaca strain T-27. Appl. Environ. Microbiol., 83, e00502–17.
Parveen, N. and Cornell, K.A. 2011. Methylthioadenosine/S-adenosylhomocystein nucleosidase, a critical enzyme for bacterial metabolism. Mol. Microbiol., 79, 7–20.
Philippot, L., Raaijmakers, J.M., Lemanceau, P., and van der Putten, W.H. 2013. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol., 11, 789–799.
Pinda, E.S., Silva, D.B., Teixeira, S.P., Coppede, J.S., Furlan, M., França, S.C., Lopes, N.P., Pereira, A.M.S., and Lopes, A.A. 2016. Mevalonate-derived quinonemethide triterpenoid from in vitro roots of Peritassa laevigata and their localization in root tissue by MALDI imaging. Sci. Rep., 6, 22627.
Price, M.N., Dehal, P.S., and Arkin, A.P. 2010. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS ONE, 5, e9490.
Rascovan, N., Carbonetto, B., Perrig, D., Díaz, M., Canciani, W., Abalo, M., Alloati, J., González-Anta, G., and Vazquez, M.P. 2016. Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci. Rep., 6, 28084.
Röttjers, L. and Faust, K. 2018. From hairballs to hypotheses-biological insights from microbial networks. FEMS Microbiol. Rev., 42, 761–780.
Salvagiotti, F., Cassman, K.G., Specht, J.E., Walters, D.T., Weiss, A., and Dobermann, A. 2008. Nitrogen uptake, fixation and response to fertilizer N in soybeans: a review. Field Crops Res., 108, 1–13.
Sang, M.K. and Kim, K.D. 2012. The volatile-producing Flavobacterium johnsoniae strain GDE09 shows biocontrol activity against Phytopthora capsici in pepper. J. Appl. Microbiol., 113, 383–398.
Segata, N., Waldron, L., Ballarini, A., Narasimhan, V., Jousson, O., and Huttenhower, C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods, 9, 811–814.
Sekowska, A., Dénervaud, V., Ashida, H., Michoud, K., Haas, D., Yokota, A., and Danchin, A. 2004. Bacterial variations on the methionine salvage pathway. BMC Microbiol., 4, 9.
Shannon, P., Markiel, A., Ozier, O., Baliga, N.S., Wang, J.T., Ramage, D., Amin, N., Schwikowski, B., and Ideker, T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504.
Spearman, C. 1904. The proof and measurement of association between two things. Am. J. Psychol., 15, 72–101.
Staib, L. and Fuchs, T.M. 2015. Regulation of fucose and 1,2-propanedioll utilization by Salmonella enterica serovar Typhimurium. Front. Microbiol., 6, 1116.
Strous, M., Fuerst, J.A., Kramer, E.H.M., Logemann, S., Muyzer, G., van de Pas-Schoonen, K.T., Webb, R., Kuenen, J.G., and Jetten, M.S.M. 1999. Missing lithotroph identified as new planctomycete. Nature, 400, 446–449.
Sugiyama, A. 2019. The soybean rhizosphere: metabolites, microbes, and beyond-a review. J. Adv. Res., 19, 67–73.
Sugiyama, A., Ueda, Y., Zushi, T., Takase, H., and Yazaki, K. 2014. Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS ONE, 9, e100709.
Tao, Y., Bu, C., Zou, L., Hu, Y., Zheng, Z., and Ouyang, J. 2021. A comprehensive review on microbial production of 1,2-proanediol: micro-organisms, metabolic pathways, and metabolic engineering. Biotechnol. Biofuels, 14, 216.
Tao, J., Meng, D., Qin, C., Liu, X., Liang, Y., Xiao, Y., Liu, Z., Gu, Y., Li, J., and Yin, H. 2018. Integrated network analysis reveals the importance of microbial interactions for maize growth. Appl. Microbiol. Biotechnol., 102, 3805–3818.
Timm, C.M., Campbell, A.G., Utturkar, S.M., Jun, S., Parales, R.E., Tan, W.A., Robeson, M.S., Lu, T.S., Jawdy, S., Brown, S.D., et al. 2015. Metabolic functions of Pseudomonas fluorescens strains from Populus deltoids depend on rhizosphere or endosphere isolation compartment. Front. Microbiol., 6, 1118.
Wang, R., Zhang, S., Ye, Y., Yu, Z., Qi, H., Zhang, H., Xue, Z., Wang, J., and Wu, M. 2019. Three new isoflavonoid glycosides from the mangrove-derived actinomycete Micromonospora aurantiaca 110B. Mar. Drugs, 17, 294.
Woese, C.R. 1987. Bacterial evolution. Microbiol. Rev., 51, 221–271.
Wotanis, C.K., Brennan, W.P. 3rd, Angotti, A.D., Villa, E.A., Zayner, J.P., Mozina, A.N., Rutkovsky, A.C., Sobe, R.C., Bond, W.G., and Karatan, E. 2017. Relative contributions of norspermidine synthesis and signaling pathways to the regulation of Vibrio cholera biofilm formation. PLoS ONE, 12, e0186291.
Zhang, B., Zhang, J., Liu, Y., Shi, P., and Wei, G. 2018. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol. Biochem., 118, 178–186.
Acknowledgements
We thank the Genomic Sequencing and Analysis Facility (GSAF) at the University of Texas at Austin for sequencing.
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of Interest
The authors have no conflict of interest to report.
Electric Supplementary Material
Rights and permissions
About this article
Cite this article
Niraula, S., Rose, M. & Chang, WS. Microbial co-occurrence network in the rhizosphere microbiome: its association with physicochemical properties and soybean yield at a regional scale. J Microbiol. 60, 986–997 (2022). https://doi.org/10.1007/s12275-022-2363-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-022-2363-x