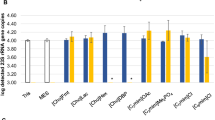
Abstract
Lipopolysaccharide (LPS) is one of the major components in the outer membrane of Gram-negative bacteria. However, its heterogeneity and variability in different bacteria and differentiation conditions make it difficult to extract all of the structural variants. We designed a solution to improve quality and biological activity of LPS extracted from various bacteria with different types of LPS, as compared to conventional methods. We introduced a quality index as a simple measure of LPS purity in terms of a degree of polysaccharide content detected by absorbance at 204 nm. Further experiments using gel electrophoresis, endotoxin test, and macrophage activation test were performed to evaluate the performance and reliability of a proposed ‘T-sol’ method and the biological effectiveness and character of the LPS products. We presented that the T-sol method had differential effects on extraction of a RAW 264.7 cell-activating LPS, which was effective in the macrophage activation with similar effects in stimulating the production of TNF-alpha. In conclusion, the T-sol method provides a simple way to improve quality and biological activity of LPS with high yield.
Similar content being viewed by others
References
Aderem, A. and Ulevitch, R.J. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406, 782–787.
Alexander, C. and Rietschel, E.T. 2001. Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 7, 167–202.
Al-Hendy, A., Toivanen, P., and Skurnik, M. 1991. Rapid method for isolation and staining of bacterial lipopolysaccharide. Microbiol. Immunol. 35, 331–333.
Baltzer, L.H. and Mattsby-Baltzer, I. 1986. Heterogeneity of lipid A: structural determination by 13C and 31P NMR of lipid A fractions from the lipopolysaccharide of Escherichia coli 0111. Biochemistry 25, 3570–3575.
Beceiro, A., Llobet, E., Aranda, J., Bengoechea, J.A., Doumith, M., Hornsey, M., Dhanji, H., Chart, H., Bou, G., Livermore, D.M., et al. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55, 3370–3379.
Bi, Y., Mann, E., Whitfield, C., and Zimmer, J. 2018. Architecture of a channel-forming O-antigen polysaccharide ABC transporter. Nature 553, 361–365.
Dai, J. and Mumper, R.J. 2010. Plant phenolics: extraction, analysis and their antioxidant and anticancer properties. Molecules 15, 7313–7352.
Darveau, R.P. and Hancock, R.W. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella Typhimurium strains. J. Bacteriol. 155, 831–838.
Davis, M.R. Jr. and Goldberg, J.B. 2012. Purification and visualization of lipopolysaccharide from Gram-negative bacteria by hot aqueous-phenol extraction. J. Vis. Exp. 63, e3916.
Dixon, D.R. and Darveau, R.P. 2005. Lipopolysaccharide heterogeneity: innate host responses to bacterial modification of lipid A structure. J. Dent. Res. 84, 584–595.
Eidhin, D.N. and Mouton, C. 1993. A rapid method for preparation of rough and smooth lipopolysaccharide from Bacteroides, Porphyromonas and Prevotella. FEMS Microbiol. Lett. 110, 133–138.
Frank, S., Specter, S., Nowotny, A., and Friedman, H. 1977. Immunocycte stimulation in vitro by nontoxic bacterial lipopolysaccharide derivatives. J. Immunol. 119, 855–860.
Funatogawa, K., Matsuura, M., Nakano, M., Kiso, M., and Hasegawa, A. 1998. Relationship of structure and biological activity of monosaccharide lipid A analogues to induction of nitric oxide production by murine macrophage RAW264.7 cells. Infect. Immun. 66, 5792–5798.
Galanos, C., Lüderitz, O., and Westphal, V.O. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9, 245–249.
Garcia-del Portillo, F., Stein, M.A., and Finlay, B.B. 1997. Release of lipopolysaccharide from intracellular compartments containing Salmonella Typhimurium to vesicles of the host epithelial cell. Infect. Immun. 65, 24–34.
Guha, M. and Mackman, N. 2001. LPS induction of gene expression in human monocytes. Cell Signal. 13, 85–94.
Henderson, J.C., O’Brien, J.P., Brodbelt, J.S., and Trent, M.S. 2013. Isolation and chemical characterization of lipid A from Gram-negative bacteria. J. Vis. Exp. 79, e50623.
Hickman, J. and Ashwell, G. 1966. Isolation of a bacterial lipopolysaccharide from Xanthomonas campestris containing 3-acetamido-3,6-dideoxy-d-galactose and d-rhamnose. J. Biol. Chem. 241, 1424–1428.
Hong, Y. and Reeves, P.R. 2014. Diversity of O-antigen repeat unit structures can account for the substantial sequence variation of Wzx translocases. J. Bacteriol. 196, 1713–1722.
Hoshino, K., Takeuchi, O., Kawai, T., Sanjo, H., Ogawa, T., Takeda, Y., Takeda, K., and Akira, S. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162, 3749–3752.
Kaijanen, L., Paakkunainen, M., Pietarinen, S., Jernström, E., and Reinikainen, S.P. 2015. Ultraviolet detection of monosaccharides: multiple wavelength strategy to evaluate results after capillary zone electrophoretic separation. Int. J. Electrochem. Sci. 10, 2950–2961.
Kalambhe, D.G., Zade, N.N., and Chaudhari, S.P. 2017. Evaluation of two different lipopolysaccharide extraction methods for purity and functionality of LPS. Int. J. Curr. Microbiol. App. Sci. 6, 1296–1302.
Kannenberg, E.L. and Carlson, R.W. 2001. Lipid A and O-chain modifications cause Rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol. Microbiol. 39, 379–392.
Kasai, N. and Nowtny, A. 1967. Endotoxic glycolipid from a heptoseless mutant of Salmonella minnesota. J. Bacteriol. 94, 1824–1836.
King, J.D., Berry, S., Clarke, B.R., Morris, R.J., and Whitfield, C. 2014. Lipopolysaccharide O antigen size distribution is determined by a chain extension complex of variable stoichiometry in Escherichia coli O9a. Proc. Natl. Acad. Sci. USA 111, 6407–6412.
Khoddami, A., Wilkes, M.A., and Roberts, T.H. 2013. Techniques for analysis of plant phenolic compounds. Molecules 18, 2328–2375.
Kobayashi, M., Utsugi, H., and Matsuda, K. 1986. Intensive UV absorption of dextrans and its application to enzyme reactions. Agric. Biol. Chem. 50, 1051–1053.
Lerouge, I. and Vanderleyden, J. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26, 17–47.
Luchi, M. and Morrison, D.C. 2000. Comparable endotoxic properties of lipopolysaccharides are manifest in diverse clinical isolates of Gram-negative bacteria. Infect. Immun. 68, 1899–1904.
Lukáčová, M., Barak, I., and Kazár, J. 2008. Role of structural variations of polysaccharide antigens in the pathogenicity of Gram-negative bacteria. Clin. Microbiol. Infect. 14, 200–206.
Luke, N.R., Sauberan, S.L., Russo, T.A., Beanan, J.M., Olson, R., Loehfelm, T.W., Cox, A.D., Michael, F.St., Vinogradov, E.V., and Campagnari, A.A. 2010. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect. Immun. 78, 2017–2023.
Maldonado, R.F., Sá-Correia, I., and Valvano, M.A. 2016. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEM Microbiol. Rev. 40, 480–493.
Marshall, J.M. and Gunn, J.S. 2015. The O-antigen capsule of Salmonella enterica serovar Typhimurium facilitates serum resistance and surface expression of FliC. Infect. Immun. 83, 3946–3959.
McAleer, J.P., Zammit, D.J., Lefrançois, L., Rossi, R.J., and Vella, A.T. 2007. The lipopolysaccharide adjuvant effect on T cells relies on nonoverlapping contributions from the MyD88 pathway and CD11c+ cells. J. Immunol. 179, 6524–6535.
Morrison, D.C. and Leive, L. 1975. Fractions of lipopolysaccharide from Escherichia coli O111:B4 prepared by two extraction procedures. J. Biol. Chem. 250, 2911–2919.
Nurminen, M. and Vaara, M. 1996. Methanol extracts LPS from deep rough bacteria. Biochem. Biophys. Res. Comm. 219, 441–444.
Pamar, A.S. and Muschol, M. 2009. Hydration and hydrodynamic interactions of lysozyme: effects of chaotropic versus kosmotropic ions. Biophys. J. 97, 590–598.
Parameswaran, N. and Patial, S. 2010. Tumor necrosis factor-α signaling in macrophages. Crit. Rev. Eukaryot. Gene Expr. 20, 87–103.
Perdomo, R. and Montero, V. 2006. Purification of E. coli 055:B5 lipopolysaccharides by size exclusion chromatography. Biotecnol. Apl. 23, 124–129.
Pier, G.B. 2007. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int. J. Med. Microbiol. 297, 277–295.
Plevin, R.E., Knoll, M., McKay, M., Arbabi, S., and Cuschieri, J. 2016. The role of lipopolysaccharide structure in monocyte activation and cytokine secretion. Shock 45, 22–27.
Poltorak, A., He, X., Smirnova, I., Liu, M.Y., Huffel, C.V., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., et al. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088.
Raetz, C.R. and Whitfield, C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700.
Raff, R.A. and Wheat, R.W. 1968. Carbohydrate composition of the phenol-soluble lipopolysaccharides of Citrobacter freundii. J. Bacteriol. 95, 2035–2043.
Rasool, O., Nnalue, N.A., and Jarstrand, C. 1992. The role of O-antigen polysaccharide in the activation of neutrophils by lipopolysaccharides of Salmonella species. Clin. Exp. Immunol. 90, 63–67.
Reuhs, B.L., Geller, D.P., Kim, J.S., Fox, J.E., Kolli, V.S.K., and Pueppke, S.G. 1998. Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl. Environ. Microbiol. 64, 4930–4938.
Rezania, S., Amirmozaffari, N., Tabarraei, B., Jeddi-Tehrani, M., Zarei, O., Alizadeh, R., Masjedian, F., and Zarnani, A.H. 2011. Extraction, purification and characterization of lipopolysaccharide from Escherichia coli and Salmonella typhi. Avicenna J. Med. Biotechnol. 3, 3–9.
Richie, D.L., Takeoka, K.T., Bojkovic, J., Metzger IV, L.E., Rath, C.M., Sawyer, W.S., Wei, J.R., and Dean, C.R. 2016. Toxic accumulation of LPS pathway intermediates underlies the requirement of LpxH for growth of Acinetobacter baumannii ATCC 19606. PLoS One 11, e0160918.
Rittig, M.G., Kaufmann, A., Robins, A., Shaw, B., Sprenger, H., Gemsa, D., Foulongne, V., Rouot, B., and Dornand, J. 2003. Smooth and rough lipopolysaccharide phenotypes of Brucella induce different intracellular trafficking and cytokine/chemokine release in human monocytes. J. Leukoc. Biol. 74, 1045–1055.
Rosenfeld, Y. and Shai, Y. 2006. Lipopolysaccharide (endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim. Biophys. Acta 1758, 1513–1522.
Sarkar, S., Ulett, G.C., Totsika, M., Phan, M.D., and Schembri, M.A. 2014. Role of capsule and O antigen in the virulence of uropathogenic Escherichia coli. PLoS One 9, e94786.
Sawyer, W.H. and Puckridge, J. 1973. The dissociation of protein by chaotropic salts. J. Biol. Chem. 248, 8429–8433.
Saxena, R.K., Vallyathan, V., and Lewis, D.M. 2003. Evidence for lipopolysaccharide-induced differentiation of RAW264.7 murine macrophage cell line into dendritic like cells. J. Biosci. 28, 129–134.
Schnaitman, C.A. and Klena, J.D. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57, 655–682.
Shimada, M., Kadowaki, T., Taniguchi, Y., Inagawa, H., Okazaki, K., and Soma, G. 2012. The involvement of O-antigen polysaccharide in lipopolysaccharide in macrophage activation. Anticancer Res. 32, 2337–2342.
Silipo, A. and Molinaro, A. 2010. The diversity of the core oligosaccharide in lipopolysaccharides, pp. 69–99. In Wang X. and Quinn P. (eds.), Endotoxins: structure, function, and recognition. subcellular biochemistry, vol 53. Springer, Dordrecht, The Netherlands.
Silipo, A., Molinaro, A., Cescutti, P., Bedini, E., Rizzo, R., Parrilli, M., and Lanzetta, R. 2005. Complete structural characterization of the lipid A fraction of a clinical strain of B. cepacia genomovar I lipopolysaccharide. Glycobiology 15, 561–570.
Sonesson, A., Jantzen, E., Tangen, T., and Zähringer, U. 1994. Chemical characterization of lipopolysaccharides from Legionella feeleii, Legionella hackeliae, and Legionella jordanis. Microbiology 140, 2663–2671.
Tirsoaga, A., Novikov, A., Adib-Conquy, M., Werts, C., Fitting, C., Cavaillon, J.M., and Caroff, M. 2007. Simple method for repurification of endotoxins for biological use. Appl. Environ. Microbiol. 73, 1803–1808.
Uchida, K. and Mizushima, S. 1987. A simple method for isolation of lipopolysaccharides from Pseudomonas aeruginosa and some other bacterial strains. Agric. Biol. Chem. 51, 3107–3114.
Wang, Z., Wang, J., Ren, G., Li, Y., and Wang, X. 2015. Influence of core oligosaccharide of lipopolysaccharide to outer membrane behavior of Escherichia coli. Mar. Drugs 13, 3325–3339.
Westphal, V.O., Lüderitz, O., and Bister, F. 1952. Über die extraktion von bakterien mit phenol/wasser. Z. Naturforschg. 7b, 148–155.
Wu, E.L., Engström, O., Jo, S., Stuhlsatz, D., Yeom, M.S., Klauda, J.B., Widmalm, G., and Im, W. 2013. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys. J. 105, 1444–1455.
Acknowledgements
This study was supported by the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (Grant number 2016R1A2B2014493 to YHK).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Nguyen, M.P., Tran, L.V.H., Namgoong, H. et al. Applications of different solvents and conditions for differential extraction of lipopolysaccharide in Gram-negative bacteria. J Microbiol. 57, 644–654 (2019). https://doi.org/10.1007/s12275-019-9116-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-019-9116-5