Abstract
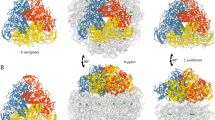
The gene product of dddC (Uniprot code G5CZI2), from the Gram-negative marine bacterium Oceanimonas doudoroffii, is a methylmalonate-semialdehyde dehydrogenase (OdoMMSDH) enzyme. MMSDH is a member of the aldehyde dehydrogenase superfamily, and it catalyzes the NADdependent decarboxylation of methylmalonate semialdehyde to propionyl-CoA. We determined the crystal structure of OdoMMSDH at 2.9 Å resolution. Among the twelve molecules in the asymmetric unit, six subunits complexed with NAD, which was carried along the protein purification steps. OdoMMSDH exists as a stable homodimer in solution; each subunit consists of three distinct domains: an NAD-binding domain, a catalytic domain, and an oligomerization domain. Computational modeling studies of the OdoMMSDH structure revealed key residues important for substrate recognition and tetrahedral intermediate stabilization. Two basic residues (Arg103 and Arg279) and six hydrophobic residues (Phe150, Met153, Val154, Trp157, Met281, and Phe449) were found to be important for tetrahedral intermediate binding. Modeling data also suggested that the backbone amide of Cys280 and the side chain amine of Asn149 function as the oxyanion hole during the enzymatic reaction. Our results provide useful insights into the substrate recognition site residues and catalytic mechanism of OdoMMSDH.
Similar content being viewed by others
References
Adams, P.D., Grosse-Kunstleve, R.W., Hung, L.W., Ioerger, T.R., McCoy, A.J., Moriarty, N.W., Read, R.J., Sacchettini, J.C., Sauter, N.K., and Terwilliger, T.C. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954.
Bannerjee, D., Sanders, L.E., and Sokatch, J.R. 1970. Properties of purified methylmalonate semialdehyde dehydrogenase of Pseudomonas aeruginosa. J. Biol. Chem. 245, 1828–1835.
Battye, T.G.G., Kontogiannis, L., Johnson, O., Powell, H.R., and Leslie, A.G. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281.
Bchini, R., Dubourg-Gerecke, H., Rahuel-Clermont, S., Aubry, A., Branlant, G., Didierjean, C., and Talfournier, F. 2012. Adenine binding mode is a key factor in triggering the early release of NADH in coenzyme A-dependent methylmalonate semialdehyde dehydrogenase. J. Biol. Chem. 287, 31095–31103.
Chen, V.B., Arendall, W.B., Headd, J.J., Keedy, D.A., Immormino, R.M., Kapral, G.J., Murray, L.W., Richardson, J.S., and Richardson, D.C. 2009. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21.
Curson, A.R., Fowler, E.K., Dickens, S., Johnston, A.W., and Todd, J.D. 2012. Multiple DMSP lyases in the γ-proteobacterium Oceanimonas doudoroffii. Biogeochemistry 110, 109–119.
Curson, A.R., Sullivan, M.J., Todd, J.D., and Johnston, A.W. 2011a. DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME J. 5, 1191–1200.
Curson, A.R., Todd, J.D., Sullivan, M.J., and Johnston, A.W. 2011b. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 9, 849–859.
Emsley, P. and Cowtan, K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132.
Hempel, J., Nicholas, H., and Lindahl, R. 1993. Aldehyde dehydrogenases: widespread structural and functional diversity within a shared framework. Protein Sci. 2, 1890–1900.
Johansson, K., Ramaswamy, S., Eklund, H., El-Ahmad, M., Hjelmqvist, L., and Jörnvall, H. 1998. Structure of betaine aldehyde dehydrogenase at 2.1 Å resolution. Protein Sci. 7, 2106–2117.
Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291.
Liu, Z.J., Sun, Y.J., Rose, J., Chung, Y.J., Hsiao, C.D., Chang, W.R., Kuo, I., Perozich, J., Lindahl, R., and Hempel, J. 1997. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struc. Mol. Biol. 4, 317–326.
Murshudov, G.N., Skubák, P., Lebedev, A.A., Pannu, N.S., Steiner, R.A., Nicholls, R.A., Winn, M.D., Long, F., and Vagin, A.A. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367.
Ni, L., Zhou, J., Hurley, T.D., and Weiner, H. 1999. Human liver mitochondrial aldehyde dehydrogenase: three-dimensional structure and the restoration of solubility and activity of chimeric forms. Protein Sci. 8, 2784–2790.
Nwachukwu, J.C., Southern, M.R., Kiefer, J.R., Afonine, P.V., Adams, P.D., Terwilliger, T.C., and Nettles, K.W. 2013. Improved crystallographic structures using extensive combinatorial refinement. Structure 21, 1923–1930.
Reisch, C.R., Moran, M.A., and Whitman, W.B. 2011. Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2, 172.
Steele, M., Lorenz, D., Hatter, K., Park, A., and Sokatch, J. 1992. Characterization of the mmsAB operon of Pseudomonas aeruginosa PAO encoding methylmalonate-semialdehyde dehydrogenase and 3-hydroxyisobutyrate dehydrogenase. J. Biol. Chem. 267, 13585–13592.
Stefels, J. 2000. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 43, 183–197.
Steinmetz, C.G., Xie, P., Weiner, H., and Hurley, T.D. 1997. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure 5, 701–711.
Stines-Chaumeil, C., Talfournier, F., and Branlant, G. 2006. Mechanistic characterization of the MSDH (methylmalonate semialdehyde dehydrogenase) from Bacillus subtilis. Biochem. J. 395, 107–115.
Sunda, W., Kieber, D., Kiene, R., and Huntsman, S. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320.
Talfournier, F., Stines-Chaumeil, C., and Branlant, G. 2011. Methylmalonate-semialdehyde dehydrogenase from Bacillus subtilis substrate specificity and coenzyme A binding. J. Biol. Chem. 286, 21971–21981.
Todd, J.D., Curson, A.R., Kirkwood, M., Sullivan, M.J., Green, R.T., and Johnston, A.W. 2011. DddQ, a novel, cupin‐containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ. Microbiol. 13, 427–438.
Todd, J.D., Curson, A.R., Nikolaidou-Katsaraidou, N., Brearley, C.A., Watmough, N.J., Chan, Y., Page, P.C., Sun, L., and Johnston, A.W. 2010. Molecular dissection of bacterial acrylate catabolism–unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ. Microbiol. 12, 327–343.
Todd, J.D., Kirkwood, M., Newton-Payne, S., and Johnston, A.W. 2012. DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 6, 223–226.
Vagin, A. and Teplyakov, A. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallog. 30, 1022–1025.
Vasiliou, V. and Nebert, D.W. 2005. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum. Genomics 2, 138.
Winn, M.D., Ballard, C.C., Cowtan, K.D., Dodson, E.J., Emsley, P., Evans, P.R., Keegan, R.M., Krissinel, E.B., Leslie, A.G., and McCoy, A. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242.
Zhang, Y.X., Tang, L., and Hutchinson, C.R. 1996. Cloning and characterization of a gene (msdA) encoding methylmalonic acid semialdehyde dehydrogenase from Streptomyces coelicolor. J. Bacteriol. 178, 490–495.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Do, H., Lee, C.W., Lee, S.G. et al. Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii . J Microbiol. 54, 114–121 (2016). https://doi.org/10.1007/s12275-016-5549-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-016-5549-2