Abstract
The complex orchestration of gene expression that mediates the transition of epithelial cells into mesenchymal cells is implicated in cancer development and metastasis. As the primary regulator of the process, epithelial-mesenchymal transition-regulating transcription factors (EMT-TFs) play key roles in metastasis. They are also highlighted in recent preclinical studies on resistance to cancer therapy. This review describes the role of three main EMT-TFs, including Snail, Twist1, and zinc-finger E homeobox-binding 1 (ZEB1), relating to drug resistance and current possible approaches for future challenges targeting EMT-TFs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metastatic cancer is the leading cause of cancer deaths worldwide, despite large-scale clinical trials by researchers to overcome it. The 5-year relative survival rate of lung cancer patients is 5%, with more than half of the patients diagnosed with metastatic disease (Howlader et al. 2015; Siegel et al. 2020). In addition, the 5-year survival rate after resection of metastatic ovarian tumors is less than 20% (Yada-Hashimoto et al. 2003). These cases emphasize that cancer metastasis plays a vital role in promoting cancer progression and reducing patient survival (Steeg 2006; Yeung et al. 2015).
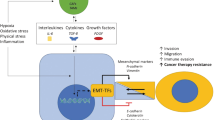
Cancer metastasis is caused by a reversible biological process, that induces the transition from non-metastatic cancer cells to metastatic cancer cells. In epithelial cancer, epithelial cells are converted into mesenchymal cells that have increased invasive properties due to loss of intercellular adhesion and thus gain of motility. The process is known as epithelial–mesenchymal transition (EMT), initially called epithelial–mesenchymal transformation (Fig. 1) (Kalluri and Neilson 2003; Kalluri and Weinberg 2009; Thiery et al. 2009; Bradley et al. 2013; Cho et al. 2019). EMT is pivotal in embryogenesis and is recognized as an essential process in cancer metastasis (Thiery 2003; Barrallo-Gimeno and Nieto 2005). Epithelial cells are characterized by apical–basal polarity, which helps cells tightly position each other on a basement membrane via intercellular junctions (Lamouille et al. 2014). Through EMT, cancer cells lose the cell–cell adhesion and gain enhanced motility and invasiveness. Several molecular processes need to be engaged during EMT initiation, such as the activation of transcription factors, expression of specific cell surface proteins and cytoskeletal proteins, production of extracellular matrix (ECM)-degrading enzymes, and changes in expression of a particular pool of microRNAs (Serrano-Gomez et al. 2016). In addition, cells that have undergone EMT often become resistant to apoptosis and senescence (Thiery et al. 2009).
Epithelial–mesenchymal transition (EMT) in cancer metastasis. In the process of cancer metastasis, the characteristics of epithelial cells change to typical mesenchymal cells. These cells have the flexibility to penetrate blood vessels. They travel along the blood vessel and then settle to develop cancer. Cancer metastasis occurs when cells from the primary cancer site travel to a secondary site. MET, mesenchymal–epithelial transition
Chemotherapy is the most widely used among various approaches to treat cancer (DeVita and Chu 2008; Min and Lee 2021). Unfortunately, drug therapies have become increasingly problematic because of resistant mechanisms in response to cancer-targeting drugs (Song and Faber 2019). Two types of drug resistance are recognized, namely intrinsic and acquired drug resistance. Intrinsic drug resistance exists in cancer cells before drug treatment, whereas acquired drug resistance develops after the exposure of cancer cells to drug treatment (Lippert et al. 2011). When multi-drug resistance occurs in cancer cells, the possibility of cancer metastasis and recurrence increases, and the clinical outcomes become worse (Hao et al. 2010). In addition, chemotherapy resistance has been observed with an increased ability to invade cells (Kajiyama et al. 2007; Işeri et al. 2011; Zhang et al. 2014). According to a recent study, the development of drug resistance leads to EMT through upregulation of EMT-promoting transcription factors in breast cancer (Mallini et al. 2014; Duran et al. 2017). Therefore, EMT has been investigated to understand the mechanism of cancer metastasis and responses to anti-cancer drugs (Oliveras-Ferraros et al. 2012; Wilson et al. 2014; Du and Shim 2016).
EMT signaling and EMT-transcription factors
The gene expression pattern during EMT is regulated indirectly or directly by EMT-transcription factors (EMT-TFs) such as Snail, Twist1, and the zinc-finger E homeobox-binding 1 (ZEB1) (Fig. 2) (Bradley et al. 2013). Subsequent EMT induction by EMT-TFs is associated with invasion, propagation, metastasis, and cancer stem cell phenotype (Thiery et al. 2009; Puisieux et al. 2014). EMT-TFs are also correlated with resistance to chemotherapy, radiation, and targeted therapy (Sequist et al. 2011; Davis et al. 2014; Ansieau et al. 2014).
Structure of EMT-related transcription factors (EMT-TFs). The size scale of each domain reflects the domain sequence length within the amino acid sequence in each protein. The N-terminal is located on the left and C-terminal is located on the right. SRD serine-rich domain, NES nuclear export signal, ZnF zinc fingers, NLS nuclear localization sequences, bHLH basic helix-loop-helix, WR tryptophan and arginine motif, SBD Smad binding Dodoma, HD homeodomain, CID CtBP interaction domain
EMT-TFs are regulated by various intracellular signaling pathways (Fig. 3a). The extracellular signal molecules bind to each specific membrane receptor and trigger intracellular signal transduction; growth factors, transforming growth factor beta (TGF-β), Wnt, and the Jagged family bind to receptor tyrosine kinase, TGF-β receptor, Frizzled, and Notch, respectively (Polyak and Weinberg 2009; Bradley et al. 2013; Lu and Kang 2019). Ligand-bound receptors transduce intracellular signals via the pathways, such as mitogen-activated protein kinase, phosphatidylinositol 3‑kinase (PI3K)/protein kinase B (Akt), nuclear factor-κB, β-catenin, or the Smad signaling pathway, which regulate the expression and stability of EMT-TFs (Bradley et al. 2013). EMT-TFs suppress the expression of epithelial markers, such as E-cadherin, cytokeratins, and tight junction proteins, and induce the expression of mesenchymal markers, including N-cadherin, vimentin, fibronectin, α-smooth muscle actin, matrix metalloproteinases (MMPs), and lethal giant larvae protein homolog 1/2 (Fig. 3b) (Kalluri and Weinberg 2009; Chao et al. 2014; Lu and Kang 2019). Reduced E-cadherin is the most representative indicator of cancer stemness and cancer resistance to treatment (Mani et al. 2008; Gupta et al. 2009). This orchestration of intracellular pathways and gene expressions regarding EMT-TFs is essential for the EMT process and the cancer therapeutic resistance correlated with EMT.
Key intracellular pathways and transcriptional target genes of EMT-TFs. a Several intracellular pathways induce transcription of EMT-TFs by binding to their promoter regions. b EMT-TFs regulate the expression of essential genes for EMT and drug-resistant related genes. NF-κB nuclear factor-κB, IKK IκB kinase, TNF-α tumor necrosis factor- α, TNFR tumor necrosis factor receptor, TNF-β transforming growth factor beta, GSK-3β glycogen synthase kinase-3β, TCF/LEF T cell factor/lymphoid enhancer factor, HIF1α hypoxia-inducible factor 1 α, IL-6 interleukin-6, JAK1/2 Janus kinase 1/2, STAT3 signal transducers and activators of transcription 3, PARP1 poly (ADP-ribose) polymerase, ERCC1 excision repair cross-complementing group 1, GAS6 growth arrest-specific 6, L1CAM L1 cell adhesion molecule, BIM Bcl-2-like protein 11, ZO-1 zonula occludens-1, ATM ataxia-telangiectasia mutated, LLGL1/2 lethal giant larvae protein homolog 1/2, α-SMA α-smooth muscle actin
In the following, we will explore how EMT-related signaling factors affect drug resistance. Epidermal growth factor receptor (EGFR) is overexpressed in human cancers, and its activation is required for TGF-β1-induced EMT (Grandis and Sok 2004; Li et al. 2015). Thomson et al. suggested that the sensitivity to inhibition of EGFR depends on the degree of EMT occurrence in EGFR-expressing human non-small cell lung cancer (NSCLC) xenograft (Thomson et al. 2005). In particular, when E-cadherin was expressed, it showed higher sensitivity to EGFR inhibition (Thomson et al. 2005). Furthermore, a decrease of E-cadherin and an increase of vimentin was observed in gefitinib-resistant A549 lung adenocarcinoma cells. These cells also revealed cross-resistance against other EGFR tyrosine kinase inhibitors (EGFR-TKI) such as erlotinib and ZD6478 (Rho et al. 2009). These studies imply that the EMT process is vital in determining sensitivity to EGFR and its inhibitors. Notably, the extent of the EMT process is essential for predicting responses to the receptor, the uppermost molecule of intracellular signaling vital for cancer development.
Akt2, which is closely related to Twist1 in the EMT process, has also been associated with cancer resistance. Akt2 is thought to be responsible for chemoresistance to docetaxel via protection of survivin in A2780 or MDA-MB-231 cells (Xing et al. 2008a, , b). Other studies suggest that growth factors induce cell growth and resistance to tamoxifen by regulating PI3K/Akt2 signaling (Sun et al. 2001). In addition, Akt2 is involved in resistance to several chemotherapies regulated by HER2 in MCF-7 cells (Zhang et al. 2011).
MMPs are essential proteolytic enzymes that promote the migration of cancer cells to adjacent tissues through ECM breakdown (Chen and Parks 2009). Invasive cancer cells have a high level of MMP-2/9 expression, and the expression level of MMP-2/9 in drug-resistant cancer cells is higher than that of drug-sensitive cancer cells (Wattanawongdon et al. 2015). A study on MMP-2/9 expression and drug resistance conducted with epirubicin revealed that the expression of MMP-2/9 has been significantly higher in epirubicin-resistant cancer cells than non-resistant cancer cells (Zhang et al. 2015).
As described above, cancer resistance and the EMT process are closely linked to each other. The signaling molecules involved in EMT mediate the resistance to drugs in various types of cancer. The effects of EMT-TFs on cancer drug resistance are not limited to the function of EMT-TFs but may occur by the regulation of related signaling factors involved. Thus, the close association between EMT-related signaling factors and drug resistance suggests that EMT-TFs may be correlated with drug resistance.
Drug resistance: Snail
Snail (Snail1) and Slug (Snail2) are reported to be associated with the EMT process during development of cancer, whereas functions of Smug (Snail3) are not revealed clearly. The Snail family shares a common C-terminal domain and various N-terminal regions (Barrallo-Gimeno and Nieto 2009). Snail proteins have multiple serine and proline residues near the middle region (Sefton et al. 1998). The C-terminal region of Snail is constructed with a DNA-binding domain that consists of zinc fingers (ZnFs) recognizing a common E2-box type element (CAGGTG) (Grimes et al. 1996; Hemavathy et al. 2000; Barrallo-Gimeno and Nieto 2009). The expression of Snail is associated with chemo- and radio-resistance through reduction of apoptosis and an increase in cell stemness (Smith and Bhowmick 2016).
Activation of P-glycoprotein (P-gp) mediates drug efflux transport, which then associates with multi-drug resistance (MDR). This activation is involved in cancer progression and is found in Snail-overexpressing NSCLC cells (Tomono et al. 2017). Overexpression of Snail influences the expression of P-gp (Tomono et al. 2017). Snail overexpression also leads to breast cancer resistance protein-mediated the MDR in MCF-7 breast cancer cell line (da Fonseca et al. 2016). Snail overexpression induces doxorubicin resistance, which subsequently causes EMT, and cells undergoing EMT show MDR through enhancing P-gp expression (Li et al. 2011). The EMT program and the induction of Snail play an acceptable role in gaining resistance to doxorubicin (Li et al. 2009). Snail-mediated upregulation of Poly [ADP-Ribose] Polymerase 1 has also been reported to contribute to doxorubicin resistance in human MDA-MB-231 breast cancer cells (Mariano et al. 2015).
Cisplatin is one of the broad and effective anti-cancer drugs (Shen et al. 2012). Co-expression of Snail and excision repair cross-complementing group 1 (ERCC1) in head and neck squamous cell carcinoma (HNSCC) patients correlated with cisplatin resistance and poor prognosis (Hsu et al. 2010). Snail knockdown induced inhibition of ERCC1 expression and attenuation of cisplatin resistance (Hsu et al. 2010). All Snail members contain a conserved SNAG domain at the N-terminus (1–9 amino acids), which is essential to repress transcription of target genes (Grimes et al. 1996; Batlle et al. 2000; Hemavathy et al. 2000; Peinado et al. 2004; Molina-Ortiz et al. 2012). The SNAG domain of Snail is a crucial mediator of ERCC1 transcription, and Snail improves sensitivity to cisplatin through inhibition of ERCC1 in HNSCC cells overexpressing Snail protein (Hsu et al. 2010). In addition, the mesenchymal phenotype is reversed and decreases resistance to cisplatin by reducing the expression of Snail in cisplatin sensitive (A2780) and resistant (A2780CR) ovarian adenocarcinoma cell lines (Haslehurst et al. 2012). These studies suggest that EMT-related transcription factors may play an essential role in cisplatin resistance in cancer. When Snail is knocked-out in cisplatin-resistant A2780CR cells, the drug sensitivity of these cells is restored, followed by reversing EMT phenotypes (Haslehurst et al. 2012).
Snail is also involved in 5-fluorouracil (5-FU) resistance. When Snail is knocked-down in MCF-7 breast cancer cells resistant to 5-FU, cell invasion is decreased through reversing EMT, and sensitivity to 5-FU is improved (Zhang et al. 2012). Considering the numerous reports regarding Snail and the correlation with chemoresistance, targeting Snail to overcome cancer resistance is an attractive approach (Fig. 4).
Drug resistance: Twist1
Twist1 was discovered originally in Drosophila as a basic helix-loop-helix family transcription factor (Simpson 1983; Thisse et al. 1987, 1988). Twist1 is reported to function as an essential transcription factor that regulates the EMT process, and eventually contributes to cancer metastasis. Twist1 is known to induce tumorigenesis in breast and prostate carcinomas, which is the result of EMT induction, invasion, and metastasis (Yang et al. 2004, 2008; Cheng et al. 2007, 2008; Pham et al. 2007; Ansieau et al. 2008; Zhuo et al. 2008). Twist1 expression induces a morphological change associated with EMT and significantly elevates cancer stem cell-like traits in MCF-7 and cervical cancer HeLa cells. Twist1 is also implicated in playing an essential role in the expansion of cancer stem cells and resistance to chemotherapy (Qin et al. 2012). Twist1 may be involved in other tumor-promoting effects, such as chemical resistance to metastasis and invasiveness associated with common chemotherapy (Li and Zhou 2011; Owens and Naylor 2013). Twist1 is not only involved in the development of acquired drug resistance in human cancer cells but is also associated with chemotherapy resistance when overexpressed, leading to a poorer prognosis (Lu et al. 2014; Deng et al. 2016; Liu et al. 2017). Interestingly, β-catenin and Akt pathways are activated in Twist1-overexpressing cells (Li and Zhou 2011).
Twist1-overexpressing breast cancer cells up-regulated Akt2 transcription, which promotes resistance to paclitaxel (Cheng et al. 2007). Similarly, resistance to paclitaxel by Twist1 overexpression occurs in human nasopharyngeal carcinoma cells, bladder, and ovarian cancer cells (Wang et al. 2004).
Erlotinib is an ATP-competitive tyrosine kinase inhibitor specific to EGFR, which is approved by the Food and Drug Administration for targeting NSCLC (Kosaka et al. 2011). Twist1 overexpression causes resistance to erlotinib in NSCLC models both in vitro and in vivo (Yochum et al. 2019). The erlotinib resistance mediated by Twist1 is due partially to the suppression of transcription of BCL2L11, a pro-apoptotic gene, by Twist1 (Yochum et al. 2019). Inhibiting Twist1 by genetic silencing or a chemical inhibitor is sufficient to overcome resistance to erlotinib (Yochum et al. 2019).
Twist1 is also suggested to be a useful biomarker for predicting resistance to doxorubicin (Li et al. 2009), used widely for adjuvant chemotherapy of breast cancer (Demir et al. 2019). TWIST1 expression level is higher in the doxorubicin-resistant samples than the doxorubicin-sensitive samples collected from breast cancer patients (Demir et al. 2019). When Twist1 is overexpressed in cisplatin and doxorubicin-resistant cancer cells, it modulates Y-box-binding protein-1 as a downstream target (Shiota et al. 2011). This correlation increases cell growth, invasion, and drug resistance (Shiota et al. 2011). Interestingly, silencing TWIST1 restores doxorubicin sensitivity in HepG2 liver cancer cells (Li et al. 2018). The positive correlation of Twist1 and multi-drug resistance protein 1 (MDR1) is observed in cancerous liver tissues, which are associated with EMT markers (Li et al. 2018). The down-regulation of Twist1 leads cancer cells to be sensitive to doxorubicin by suppression of MDR1 and EMT (Li et al. 2018). Chemotherapy increases the expression of Twist1 and several ATP-binding cassette transporters in invasive cancer cells, but not in non-invasive cells (Saxena et al. 2011). Chen et al. have shown that the expression of Twist1 causes resistance to an anthracycline, a class of drugs including doxorubicin and epirubicin, by regulating the expression of P-gp in the bladder cancer cell line (Chen et al. 2012).
In vivo tumorigenic assays show that Twist1-overexpressing cells, which are resistant to cisplatin, result in the widespread dissemination of tumors lining the peritoneal cavity walls (Roberts et al. 2016). Twist1 is believed to drive cisplatin resistance via upregulation of L1CAM, GAS6, and Akt signaling pathways in an ovarian cancer model (Roberts et al. 2016). Suppression of Twist1 expression by introducing miR-186 renders the ovarian cancer cells to overcome cisplatin resistance (Zhu et al. 2016).
Because the high expression level of Twist1 affects the responses to various chemotherapies in different types of cancer, the proper modulation or genetic ablation of Twist1 needs to be studied for the further possibility to restore sensitivity to the cancer drugs.
Drug resistance: ZEB1
The ZEB transcription factor family consists of ZEB1 (also named TCF8) and ZEB2 (also known as SIPI) (Anose and Sanders 2011). ZEB family proteins contain two ZnF domains, N-terminal ZnF and C-terminal ZnF. The helix-loop-helix motif of ZEB allows for binding to the E-box within the E-cadherin promoter region with high specificity (Peinado et al. 2007). Phosphorylation within C-terminal ZnF inhibits the binding of ZEB to DNA and its transcriptional activity (Llorens et al. 2016).
ZEB1 is correlated with resistance to various drugs in many types of cancer (Witta et al. 2006; Arumugam et al. 2009; Tryndyak et al. 2010; Chang et al. 2011). MDR1 reduces the intracellular accumulation of drugs such as cisplatin, 5-FU, and doxorubicin, by functioning as an ATP-dependent drug outflow pump (Janigro et al. 2006; McCormick et al. 2015). Yoshida et al. demonstrated that ZEB1 overexpression acquires the resistance of EGFR-TKI in human NSCLC samples and cells (Yoshida et al. 2016).
Pancreatic cancer cells show different sensitivity to multiple chemotherapeutic drugs, such as gemcitabine, 5-FU, and cisplatin; MIAPaCa-2, PANC-1, Hs766T, AsPC-1, and Mpanc96 cell lines are resistant, whereas CFPAC-1, L3.6pl, BxPC-3, and SU86.86 are sensitive to the drugs (Arumugam et al. 2009). The two distinct groups of cell lines differ in the expression pattern of EMT-related genes, mainly ZEB1. Knockdown of ZEB1 restores drug sensitivity and increases the expression of epithelial markers in cancer cells showing mesenchymal characteristics (Arumugam et al. 2009).
A colon cancer cell line with oxaliplatin (OXA) resistance, HCT116/OXA, shows a high expression level of ZEB1 along with mesenchymal markers, including vimentin, MMP-2, and MMP-9 (Guo et al. 2017). Silencing of ZEB1 restores sensitivity to OXA in HCT116/OXA, and the inverse correlation between ZEB1 expression level and OXA sensitivity occurs both in vitro and in vivo (Guo et al. 2017). In addition, ZEB1 expression level is higher in histone deacetylase inhibitor (HDACi) butyrate-resistant colorectal cancer cells (Lazarova and Bordonaro 2017). These cancer cells lack the expression of histone acetyltransferase p300, which associates with β-catenin and mediates the transcriptional activity of the Wnt signaling pathway (Lazarova and Bordonaro 2017). Interestingly, mutations in the Wnt signaling pathway are a major initiating event in colorectal cancer and regulate ZEB1 expression (Sanchez-Tillo et al. 2013). Therefore, ZEB1 expression is correlated with both cancer aggressiveness and with the responses of cancer to butyrate.
Axl as a receptor tyrosine kinase is activated by vitamin K‐dependent protein, namely growth arrest-specific protein 6 (GAS6) (Zhu et al. 2019). GAS6/Axl signaling pathway drives the survival, proliferation and invasion of cancer cells (Zhu et al. 2019). Moreover, Axl overexpression has been reported to be associated with poor prognosis in a wide range of cancers and to play an essential role in metastasis (Dunne et al. 2014; Lee et al. 2014; Reichl et al. 2015; Brand et al. 2015; Hattori et al. 2016). According to the study, Axl has a correlation with the downstream targets of breast cancer specimens, and breast cancer prognosis is not good (Wang et al. 2016). Wang et al. showed that the Akt/GSK-3β/β-catenin cascade induced transcription of ZEB1, which resulted in doxorubicin resistance.
Ataxia-telangiectasia mutated (ATM) kinase plays a key role in the homologous recombination repair of damaged DNA (So et al. 2009). Zhang et al. reported that ZEB1 induces resistance to epirubicin with increasing expression of ATM kinase (Zhang et al. 2018). They also found that the expression of ZEB1 in chemo-resistant breast cancer is significantly higher than in chemo-sensitive breast cancer (Zhang et al. 2018). Interestingly, their report shows that increased expression of ATM kinase by ZEB1 enhances DNA repair in response to the epirubicin-induced DNA breakage and thus confers resistance to epirubicin (Zhang et al. 2018).
The other resistant mechanism related to ZEB1 includes long non-coding RNA of ZEB1, ZEB1-antisense 1 (ZEB1-AS1). ZEB1-AS1 is expressed from the promoter region of ZEB1 and regulates ZEB1 expression positively (Su et al. 2017). The dysregulation of ZEB1-AS1 plays a pivotal function in tumorigenesis and tumor development (Zhao et al. 2019). Li et al. have shown that ZEB1-AS1 is upregulated in hepatocellular carcinoma and breast cancer cells and promotes metastasis (Li et al. 2016). ZEB1-AS1 regulates ZEB1 expression by binding to miR-129-5p competitively. Silenced ZEB1-AS1 inhibits resistance to cisplatin and promotes apoptosis in MCF-7 cells via upregulation of miR-129-5p and downregulation of ZEB1, Bcl-2, MDR1, and P-gp (Gao et al. 2020). The implication that ZEB1 plays a critical role in drug resistance is growing as more cases are proposed. For restoring sensitivity to cancer therapies, the appropriate approaches regulating EMT-TFs, including Snail, Twist1, and ZEB1, are required. The following section describes several strategies targeting these EMT-TFs.
Targeting EMT-TFs to suppress cancer development and overcome drug resistance
Because the EMT process and its transcription factors drive drug resistance, the inhibitors of the process and factors will be beneficial for chemotherapy or targeted therapy (Du and Shim 2016).
In mutant K-RAS-driven cancer cells, Snail binds to the DNA-binding domain of p53 and prevents p53 from DNA binding and further apoptosis or senescence induced by p53 activity (Lee et al. 2009). Although a few inhibitors are developed to bind and inhibit Snail directly, GN25 and GN29 bind Snail and inhibit this interaction between Snail and p53 (Lee et al. 2010). The K-RAS mutated-cancer cells treated with the compounds show reduced proliferation and tumor progression. Unfortunately, these compounds have a limitation in that they are only effective in K-RAS-driven cancer cells harboring wild-type p53.
Recently, Hong et al. identified a small-molecule CYD19 that binds Snail with a high affinity and inhibits the interaction of Snail with CREB-binding protein (CBP)/p300 (Li and Balazsi 2018). The disrupted interaction leads to impairment of CBP/p300-mediated Snail acetylation and accelerated degradation of Snail via the proteasomal pathway. Snail-dependent EMT, cancer stem cell expansion, and metastasis are inhibited in vivo as Snail degradation is induced by CYD19.
Given that Snail binds to the consensus E-box sequence, Vistain et al. utilized an amine-modified E-box oligonucleotide conjugated to the cobalt(III) complex to generate CoIII-Ebox (Vistain et al. 2015). As CoIII-E-box is introduced into the cells, Snail binds to CoIII-E-box, and the endogenous E-cadherin gene expression is no longer repressed by Snail. CoIII-E-box blocks the binding of Snail to the promoter regions of its target genes, thus further inhibiting EMT in breast cancer cells.
A few direct inhibitors of Snail target the interaction of Snail with its binding partners as described above. The other approach to inhibit Snail is to modulate the regulatory mechanisms of Snail. For instance, the stability of Snail can be enhanced by Dub3, a deubiquitinase of Snail. Dub3-mediated Snail stabilization is disrupted by a Dub3-specific inhibitor, WP1130, and Snail-driven metastasis is inhibited in breast cancer (Wu et al. 2017).
Harmine, a harmala alkaloid, is as a novel Twist1 inhibitor that successfully inhibits cell dissemination, growth of invadopodia in 3D culture, and proliferation of NSCLC cells (Yochum et al. 2017; Zhang et al. 2020). Harmine induces the degradation of Twist1 heterodimerized with E2A protein, a dimer partner of Twist1 (Yochum et al. 2017). In addition, harmine shows anti-cancer activity via Twist1 suppression in K-RAS mutant NSCLC mouse models (Yochum et al. 2017).
ABT-263 is an orally available analog that binds and inhibits anti-apoptotic Bcl-2/Bcl-XL. Furthermore, ZEB1 suppresses transcription of pro-apoptotic BCL2L11, which results in resistance to ABT-263. However, FK228 (also romidepsin), known as HDACi, attenuated ZEB1-induced ABT263 resistance by up-regulating Bcl-2-like protein 11 expression (Inoue-Yamauchi and Oda 2020).
The underlying mechanism of EMT-related resistance to doxorubicin is also highly dependent on miRNA expression and activity. A study by Chen et al. shows that the expression of miR-200 is reduced significantly when MCF-7 cells become resistant to doxorubicin (Chen et al. 2013). This miRNA targets the mRNA of ZEB1/2 directly (Park et al. 2008). Ectopic expression of miR-200 induces E-cadherin upregulation, vimentin downregulation, and decreased motility.
Concluding remarks
Snail, Twist, and ZEB families not only induce EMT but also are correlated with drug resistance, all of which makes cancer therapy more challenging. We have described the cancer drug resistance regarding EMT-TFs and targeting strategies of EMT-TFs. Although the strategies targeting the EMT-TFs directly are not in the clinical trials yet, the relevance of the EMT-TFs in metastatic cancer is significant in vitro, in vivo, and in clinical data. Investigation into the regulatory mechanisms of the EMT-TFs in correlation with cancer therapeutic resistance will develop novel targeting strategies towards the EMT-TFs. Eventually, different approaches to understanding the function and regulation of EMT-TFs will shed light on metastatic cancer therapy.
References
Anose BM, Sanders MM (2011) Androgen receptor regulates transcription of the ZEB1 transcription factor. Int J Endocrinol 2011:1–10. https://doi.org/10.1155/2011/903918
Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, Maestro R, Voeltzel T, Selmi A, Valsesia-Wittmann S, Caron de Fromentel C, Puisieux A (2008) Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 14:79–89. https://doi.org/10.1016/j.ccr.2008.06.005
Ansieau S, Collin G, Hill L (2014) EMT or EMT-promoting transcription factors, where to focus the light? Front Oncol 4:353. https://doi.org/10.3389/fonc.2014.00353
Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ, Choi W (2009) Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res 69:5820–5828. https://doi.org/10.1158/0008-5472.CAN-08-2819
Barrallo-Gimeno A, Nieto MA (2005) The snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132:3151–3161. https://doi.org/10.1242/dev.01907
Barrallo-Gimeno A, Nieto MA (2009) Evolutionary history of the snail/scratch superfamily. Trends Genet 25:248–252. https://doi.org/10.1016/j.tig.2009.04.001
Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, De Herreros AG (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2:84–89. https://doi.org/10.1038/35000034
Bradley CK, Norton CR, Chen Y, Han X, Booth CJ, Yoon JK, Krebs LT, Gridley T (2013) The snail family gene Snai3 is not essential for embryogenesis in mice. PLoS ONE 8:3–8. https://doi.org/10.1371/journal.pone.0065344
Brand TM, Iida M, Stein AP, Corrigan KL, Braverman CM, Coan JP, Pearson HE, Bahrar H, Fowler TL, Bednarz BP, Saha S, Yang D, Gill PS, Lingen MW, Saloura V, Villaflor VM, Salgia R, Kimple RJ, Wheeler DL (2015) AXL is a logical molecular target in head and neck squamous cell carcinoma. Clin Cancer Res 21:2601–2612. https://doi.org/10.1158/1078-0432.CCR-14-2648
Chang T-H, Tsai M-F, Su K-Y, Wu S-G, Huang C-P, Yu S-L, Yu Y-L, Lan C-C, Yang C-H, Lin S-B, Wu C-P, Shih J-Y, Yang P-C (2011) Slug confers resistance to the epidermal growth factor receptor tyrosine kinase inhibitor. Am J Respir Crit Care Med 183:1071–1079. https://doi.org/10.1164/rccm.201009-1440OC
Chao CH, Chang CC, Wu MJ, Ko HW, Wang D, Hung MC, Yang JY, Chang CJ (2014) MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest. https://doi.org/10.1172/JCI73351
Chen P, Parks WC (2009) Role of matrix metalloproteinases in epithelial migration. J Cell Biochem 108:1233–1243. https://doi.org/10.1002/jcb.22363
Chen Y, Li L, Zeng J, Wu K, Zhou J, Guo P, Zhang D, Xue Y, Liang L, Wang X, Chang LS, He D (2012) Twist confers chemoresistance to anthracyclines in bladder cancer through upregulating P-glycoprotein. Chemotherapy 58:264–272. https://doi.org/10.1159/000341860
Chen Y, Sun Y, Chen L, Xu X, Zhang X, Wang B, Min L, Liu W (2013) miRNA-200c increases the sensitivity of breast cancer cells to doxorubicin through the suppression of E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep 7:1579–1584. https://doi.org/10.3892/mmr.2013.1403
Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH (2007) Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res 67:1979–1987. https://doi.org/10.1158/0008-5472.CAN-06-1479
Cheng GZ, Zhang WZ, Sun M, Wang Q, Coppola D, Mansour M, Xu LM, Costanzo C, Cheng JQ, Wang LH (2008) Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J Biol Chem 283:14665–14673. https://doi.org/10.1074/jbc.M707429200
Cho ES, Kang HE, Kim NH, Yook JI (2019) Therapeutic implications of cancer epithelial-mesenchymal transition (EMT). Arch Pharm Res 42:14–24. https://doi.org/10.1007/s12272-018-01108-7
da Fonseca LM, da Silva VA, Freire-de-Lima L, Previato JO, Mendonça-Previato L, Capella MAM (2016) Glycosylation in cancer: interplay between multidrug resistance and epithelial-to-mesenchymal transition? Front Oncol 6:158. https://doi.org/10.3389/fonc.2016.00158
Davis FM, Stewart TA, Thompson EW, Monteith GR (2014) Targeting EMT in cancer: opportunities for pharmacological intervention. Trends Pharmacol Sci 35:479–488. https://doi.org/10.1016/j.tips.2014.06.006
Demir S, Müslümanoğlu M, Müslümanoğlu M, Başaran S, Çalay Z, Aydıner A, Vogt U, Çakır T, Kadıoğlu H, Artan S (2019) TWIST1 gene expression as a biomarker for predicting primary doxorubicin resistance in breast cancer. Balk J Med Genet 22:25–30. https://doi.org/10.2478/bjmg-2019-0025
Deng J-J, Zhang W, Xu X-M, Zhang F, Tao W-P, Ye J-J, Ge W (2016) Twist mediates an aggressive phenotype in human colorectal cancer cells. Int J Oncol 48:1117–1124. https://doi.org/10.3892/ijo.2016.3342
DeVita VT, Chu E (2008) A history of cancer chemotherapy. Cancer Res 68:8643–8653. https://doi.org/10.1158/0008-5472.CAN-07-6611
Du B, Shim J (2016) Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules 21:965. https://doi.org/10.3390/molecules21070965
Dunne PD, McArt DG, Blayney JK, Kalimutho M, Greer S, Wang T, Srivastava S, Ong CW, Arthur K, Loughrey M, Redmond K, Longley DB, Salto-Tellez M, Johnston PG, Van Schaeybroeck S (2014) AXL is a key regulator of inherent and chemotherapy-induced invasion and predicts a poor clinical outcome in early-stage colon cancer. Clin Cancer Res 20:164–175. https://doi.org/10.1158/1078-0432.CCR-13-1354
Duran GE, Wang YC, Moisan F, Francisco EB, Sikic BI (2017) Decreased levels of baseline and drug-induced tubulin polymerisation are hallmarks of resistance to taxanes in ovarian cancer cells and are associated with epithelial-to-mesenchymal transition. Br J Cancer 116:1318–1328. https://doi.org/10.1038/bjc.2017.102
Gao J, Yuan Y, Zhang L, Yu S, Lu J, Feng J, Hu S (2020) Inhibition of ZEB1-AS1 confers cisplatin sensitivity in breast cancer by promoting microRNA-129-5p-dependent ZEB1 downregulation. Cancer Cell Int 20:90. https://doi.org/10.1186/s12935-020-1164-8
Grandis JR, Sok JC (2004) Signaling through the epidermal growth factor receptor during the development of malignancy. Pharmacol Ther 102:37–46. https://doi.org/10.1016/j.pharmthera.2004.01.002
Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN (1996) The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol 16:6263–6272. https://doi.org/10.1128/mcb.16.11.6263
Guo C, Ma J, Deng G, Qu Y, Yin L, Li Y, Han Y, Cai C, Shen H, Zeng S (2017) ZEB1 promotes oxaliplatin resistance through the induction of epithelial: mesenchymal transition in colon cancer cells. J Cancer 8:3555–3566. https://doi.org/10.7150/jca.20952
Gupta PB, Chaffer CL, Weinberg RA (2009) Cancer stem cells: mirage or reality? Nat Med 15:1010–1012. https://doi.org/10.1038/nm0909-1010
Hao J, Chen H, Madigan MC, Cozzi PJ, Beretov J, Xiao W, Delprado WJ, Russell PJ, Li Y (2010) Co-expression of CD147 (EMMPRIN), CD44v3-10, MDR1 and monocarboxylate transporters is associated with prostate cancer drug resistance and progression. Br J Cancer 103:1008–1018. https://doi.org/10.1038/sj.bjc.6605839
Haslehurst AM, Koti M, Dharsee M, Nuin P, Evans K, Geraci J, Childs T, Chen J, Li J, Weberpals J, Davey S, Squire J, Park PC, Feilotter H (2012) EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 12:91. https://doi.org/10.1186/1471-2407-12-91
Hattori S, Kikuchi E, Kosaka T, Miyazaki Y, Tanaka N, Miyajima A, Mikami S, Oya M (2016) Relationship between increased expression of the Axl/Gas6 signal cascade and prognosis of patients with upper tract urothelial carcinoma. Ann Surg Oncol 23:663–670. https://doi.org/10.1245/s10434-015-4848-x
Hemavathy K, Ashraf SI, Ip YT (2000) Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene 257:1–12. https://doi.org/10.1016/S0378-1119(00)00371-1
Howlader N, Noone A, Krapcho M, Miller D, Bishop K, Altekruse S, Kosary C, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis D, Chen H, Feuer E, Cronin K (2015) SEER Cancer Statistics Review, 1975-2014, National Cancer Institute. Bethesda, MD. https://seer.cancer.gov/csr/1975_2014/
Hsu DSS, Lan HY, Huang CH, Tai SK, Chang SY, Tsai TL, Chang CC, Tzeng CH, Wu KJ, Kao JY, Yang MH (2010) Regulation of excision repair cross-complementation group 1 by snail contributes to cisplatin resistance in head and neck cancer. Clin Cancer Res 16:4561–4571. https://doi.org/10.1158/1078-0432.CCR-10-0593
Inoue-Yamauchi A, Oda H (2020) EMT-inducing transcription factor ZEB1-associated resistance to the BCL-2/BCL-XL inhibitor is overcome by BIM upregulation in ovarian clear cell carcinoma cells. Biochem Biophys Res Commun 526:612–617. https://doi.org/10.1016/j.bbrc.2020.03.139
Işeri ÖD, Kars MD, Arpaci F, Atalay C, Pak I, Gündüz U (2011) Drug resistant MCF-7 cells exhibit epithelial-mesenchymal transition gene expression pattern. Biomed Pharmacother 65:40–45. https://doi.org/10.1016/j.biopha.2010.10.004
Janigro D, Perju C, Fazio V, Hallene K, Dini G, Agarwal MK, Cucullo L (2006) Alternating current electrical stimulation enhanced chemotherapy: a novel strategy to bypass multidrug resistance in tumor cells. BMC Cancer 6:72. https://doi.org/10.1186/1471-2407-6-72
Kajiyama H, Shibata K, Terauchi M, Yamashita M, Ino K, Nawa A, Kikkawa F (2007) Chemoresistance to paclitaxel induces epithelial-mesenchymal transition and enhances metastatic potential for epithelial ovarian carcinoma cells. Int J Oncol 31:277–283. https://doi.org/10.3892/ijo.31.2.277
Kalluri R, Neilson EG (2003) Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest 112:1776–1784. https://doi.org/10.1172/JCI200320530
Kalluri R, Weinberg RA (2009) The basics of epithelial-mesenchymal transition. J Clin Invest 119:1420–1428. https://doi.org/10.1172/JCI39104
Kosaka T, Yamaki E, Mogi A, Kuwano H (2011) Mechanisms of resistance to EGFR TKIs and development of a new generation of drugs in non-small-cell lung cancer. J Biomed Biotechnol 2011:1–7. https://doi.org/10.1155/2011/165214
Lamouille S, Xu J, Derynck R (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196. https://doi.org/10.1038/nrm3758
Lazarova D, Bordonaro M (2017) ZEB1 mediates drug resistance and EMT in p300-deficient CRC. J Cancer 8:1453–1459. https://doi.org/10.7150/jca.18762
Lee H-J, Jeng Y-M, Chen Y-L, Chung L, Yuan R-H (2014) Gas6/Axl pathway promotes tumor invasion through the transcriptional activation of slug in hepatocellular carcinoma. Carcinogenesis 35:769–775. https://doi.org/10.1093/carcin/bgt372
Lee SH, Lee SJ, Yeon SJ, Xu Y, Ho SK, Ha NC, Park BJ (2009) Blocking of p53-snail binding, promoted by oncogenic K-Ras, recovers p53 expression and function. Neoplasia 11:22–31. https://doi.org/10.1593/neo.81006
Lee SH, Shen GN, Jung YS, Lee SJ, Chung JY, Kim HS, Xu Y, Choi Y, Lee JW, Ha NC, Song GY, Park BJ (2010) Antitumor effect of novel small chemical inhibitors of Snail-p53 binding in K-Ras-mutated cancer cells. Oncogene 29:4576–4587. https://doi.org/10.1038/onc.2010.208
Li C, Balazsi G (2018) A landscape view on the interplay between EMT and cancer metastasis. NPJ Syst Biol Appl 4:34. https://doi.org/10.1038/s41540-018-0068-x
Li J, Zhou BP (2011) Activation of β-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 11:49. https://doi.org/10.1186/1471-2407-11-49
Li L, Qi L, Liang Z, Song W, Liu Y, Wang Y, Sun B, Zhang B, Cao W (2015) Transforming growth factor-β1 induces EMT by the transactivation of epidermal growth factor signaling through HA/CD44 in lung and breast cancer cells. Int J Mol Med 36:113–122. https://doi.org/10.3892/ijmm.2015.2222
Li Q-Q, Xu J-D, Wang W-J, Cao X-X, Chen Q, Tang F, Chen Z-Q, Liu X-P, Xu Z-D (2009) Twist1-mediated adriamycin-induced epithelial-mesenchymal transition relates to multidrug resistance and invasive potential in breast cancer cells. Clin Cancer Res 15:2657–2665. https://doi.org/10.1158/1078-0432.CCR-08-2372
Li R, Wu C, Liang H, Zhao Y, Lin C, Zhang X, Ye C (2018) Knockdown of TWIST enhances the cytotoxicity of chemotherapeutic drugs in doxorubicin-resistant HepG2 cells by suppressing MDR1 and EMT. Int J Oncol. https://doi.org/10.3892/ijo.2018.4495
Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z (2016) Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene 35:1575–1584. https://doi.org/10.1038/onc.2015.223
Li W, Liu C, Tang Y, Li H, Zhou F, Lv S (2011) Overexpression of snail accelerates adriamycin induction of multidrug resistance in breast cancer cells. Asian Pacific J Cancer Prev 12:2575–2580
Lippert T, Ruoff H-J, Volm M (2011) Intrinsic and acquired drug resistance in malignant tumors. Arzneimittelforschung 58:261–264. https://doi.org/10.1055/s-0031-1296504
Liu Y-R, Liang L, Zhao JM, Zhang Y, Zhang M, Zhong W-L, Zhang Q, Wei J-J, Li M, Yuan J, Chen S, Zong S-M, Liu H-J, Meng J, Qin Y, Sun B, Yang L, Zhou H-G, Sun T, Yang C (2017) Twist1 confers multidrug resistance in colon cancer through upregulation of ATP-binding cassette transporters. Oncotarget 8:52901–52912. https://doi.org/10.18632/oncotarget.17548
Llorens MC, Lorenzatti G, Cavallo NL, Vaglienti MV, Perrone AP, Carenbauer AL, Darling DS, Cabanillas AM (2016) Phosphorylation regulates functions of ZEB1 transcription factor. J Cell Physiol 231:2205–2217. https://doi.org/10.1002/jcp.25338
Lu S, Yu L, Mu Y, Ma J, Tian J, Xu W, Wang H (2014) Role and mechanism of Twist1 in modulating the chemosensitivity of FaDu cells. Mol Med Rep 10:53–60. https://doi.org/10.3892/mmr.2014.2212
Lu W, Kang Y (2019) Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell 49:361–374. https://doi.org/10.1016/j.devcel.2019.04.010
Mallini P, Lennard T, Kirby J, Meeson A (2014) Epithelial-to-mesenchymal transition: what is the impact on breast cancer stem cells and drug resistance. Cancer Treat Rev 40:341–348. https://doi.org/10.1016/j.ctrv.2013.09.008
Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715. https://doi.org/10.1016/j.cell.2008.03.027
Mariano G, Ricciardi MR, Trisciuoglio D, Zampieri M, Ciccarone F, Guastafierro T, Calabrese R, Valentini E, Tafuri A, Del Bufalo D, Caiafa P, Reale A (2015) PARP inhibitor ABT-888 affects response of MDA-MB-231 cells to doxorubicin treatment, targeting snail expression. Oncotarget 6:15008–15021. https://doi.org/10.18632/oncotarget.3634
McCormick JW, Vogel PD, Wise JG (2015) Multiple drug transport pathways through human P-glycoprotein. Biochemistry 54:4374–4390. https://doi.org/10.1021/acs.biochem.5b00018
Min H-Y, Lee H-Y (2021) Mechanisms of resistance to chemotherapy in non-small cell lung cancer. Arch Pharm Res 44:146–164. https://doi.org/10.1007/s12272-021-01312-y
Molina-Ortiz P, Villarejo A, MacPherson M, Santos V, Montes A, Souchelnytskyi S, Portillo F, Cano A (2012) Characterization of the SNAG and SLUG domains of Snail2 in the repression of E-cadherin and EMT induction: modulation by serine 4 phosphorylation. PLoS ONE 7:e36132. https://doi.org/10.1371/journal.pone.0036132
Oliveras-Ferraros C, Corominas-Faja B, Cufí S, Vazquez-Martin A, Martin-Castillo B, Iglesias JM, López-Bonet E, Martin ÁG, Menendez JA (2012) Epithelial-to-mesenchymal transition (EMT) confers primary resistance to trastuzumab (Herceptin). Cell Cycle 11:4020–4032. https://doi.org/10.4161/cc.22225
Owens TW, Naylor MJ (2013) Breast cancer stem cells. Front Physiol 4:225. https://doi.org/10.3389/fphys.2013.00225
Park S-M, Gaur AB, Lengyel E, Peter ME (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22:894–907. https://doi.org/10.1101/gad.1640608
Peinado H, Ballestar E, Esteller M, Cano A (2004) Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 24:306–319. https://doi.org/10.1128/mcb.24.1.306-319.2004
Peinado H, Olmeda D, Cano A (2007) Snail, ZEB and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7:415–428. https://doi.org/10.1038/nrc2131
Pham CG, Bubici C, Zazzeroni F, Knabb JR, Papa S, Kuntzen C, Franzoso G (2007) Upregulation of Twist-1 by NF-κB blocks cytotoxicity induced by chemotherapeutic drugs. Mol Cell Biol 27:3920–3935. https://doi.org/10.1128/mcb.01219-06
Polyak K, Weinberg RA (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9:265–273. https://doi.org/10.1038/nrc2620
Puisieux A, Brabletz T, Caramel J (2014) Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol 16:488–494. https://doi.org/10.1038/ncb2976
Qin Q, Xu Y, He T, Qin C, Xu J (2012) Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res 22:90–106. https://doi.org/10.1038/cr.2011.144
Reichl P, Dengler M, van Zijl F, Huber H, Führlinger G, Reichel C, Sieghart W, Peck-Radosavljevic M, Grubinger M, Mikulits W (2015) Axl activates autocrine transforming growth factor-β signaling in hepatocellular carcinoma. Hepatology 61:930–941. https://doi.org/10.1002/hep.27492
Rho JK, Choi YJ, Lee JK, Ryoo B-Y, Il NI, Yang SH, Kim CH, Lee JC (2009) Epithelial to mesenchymal transition derived from repeated exposure to gefitinib determines the sensitivity to EGFR inhibitors in A549, a non-small cell lung cancer cell line. Lung Cancer 63:219–226. https://doi.org/10.1016/j.lungcan.2008.05.017
Roberts CM, Tran MA, Pitruzzello MC, Wen W, Loeza J, Dellinger TH, Mor G, Glackin CA (2016) TWIST1 drives cisplatin resistance and cell survival in an ovarian cancer model, via upregulation of GAS6, L1CAM, and Akt signalling. Sci Rep 6:37652. https://doi.org/10.1038/srep37652
Sanchez-Tillo E, de Barrios O, Siles L, Amendola PG, Darling DS, Cuatrecasas M, Castells A, Postigo A (2013) ZEB1 promotes invasiveness of colorectal carcinoma cells through the opposing regulation of uPA and PAI-1. Clin Cancer Res 19:1071–1082. https://doi.org/10.1158/1078-0432.CCR-12-2675
Saxena M, Stephens MA, Pathak H, Rangarajan A (2011) Transcription factors that mediate epithelial–mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell Death Dis 2:e179–e179. https://doi.org/10.1038/cddis.2011.61
Sefton M, Sánchez S, Nieto MA (1998) Conserved and divergent roles for members of the snail family of transcription factors in the chick and mouse embryo. Development 125:3111–3121. https://doi.org/10.13039/501100000780
Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, Akhavanfard S, Heist RS, Temel J, Christensen JG, Wain JC, Lynch TJ, Vernovsky K, Mark EJ, Lanuti M, Iafrate AJ, Mino-Kenudson M, Engelman JA (2011) Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3:7526. https://doi.org/10.1126/scitranslmed.3002003
Serrano-Gomez SJ, Maziveyi M, Alahari SK (2016) Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer 15:18. https://doi.org/10.1186/s12943-016-0502-x
Shen DW, Pouliot LM, Hall MD, Gottesman MM (2012) Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol Rev 64:706–721. https://doi.org/10.1124/pr.111.005637
Shiota M, Yokomizo A, Itsumi M, Uchiumi T, Tada Y, Song Y, Kashiwagi E, Masubuchi D, Naito S (2011) Twist1 and Y-box-binding protein-1 promote malignant potential in bladder cancer cells. BJU Int 108:E142–E149. https://doi.org/10.1111/j.1464-410X.2010.09810.x
Siegel RL, Miller KD, Jemal A (2020) Cancer statistics, 2020. CA Cancer J Clin 70:7–30. https://doi.org/10.3322/caac.21590
Simpson P (1983) Maternal-zygotic gene interactions during formation of the dorsoventral pattern in drosophila embryos. Genetics 105:615–632
Smith B, Bhowmick N (2016) Role of EMT in metastasis and therapy resistance. J Clin Med 5:17. https://doi.org/10.3390/jcm5020017
So S, Davis AJ, Chen DJ (2009) Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J Cell Biol. https://doi.org/10.1083/jcb.200906064
Song K-A, Faber AC (2019) Epithelial-to-mesenchymal transition and drug resistance: transitioning away from death. J Thorac Dis 11:E82–E85. https://doi.org/10.21037/jtd.2019.06.11
Steeg PS (2006) Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 12:895–904. https://doi.org/10.1038/nm1469
Su W, Xu M, Chen X, Chen N, Gong J, Nie L, Li L, Li X, Zhang M, Zhou Q (2017) Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. https://doi.org/10.1186/s12943-017-0711-y
Sun M, Paciga JE, Feldman RI, Yuan ZQ, Coppola D, You Yong Lu, Shelley SA, Nicosia SV, Cheng JQ (2001) Phosphatidylinositol-3-OH kinase (PI3K)/AKT2, activated in breast cancer, regulates and is induced by estrogen receptor α (ERα) via interaction between ERα and PI3K. Cancer Res 61:5985–8991
Thiery JP (2003) Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol 15:740–746. https://doi.org/10.1016/j.ceb.2003.10.006
Thiery JP, Acloque H, Huang RYJ, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890. https://doi.org/10.1016/j.cell.2009.11.007
Thisse B, El MM, Perrin-Schmitt F (1987) The twist gene: isolation of a drosophila zygotle gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res 15:3439–3453. https://doi.org/10.1093/nar/15.8.3439
Thisse B, Stoetzel C, Gorostiza-Thisse C, Perrin-Schmitt F (1988) Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early drosophila embryos. EMBO J 7:2175–2183. https://doi.org/10.1002/j.1460-2075.1988.tb03056.x
Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, Iwata KK, Gibson N, Haley JD (2005) Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res 65:9455–9462. https://doi.org/10.1158/0008-5472.CAN-05-1058
Tomono T, Yano K, Ogihara T (2017) Snail-induced epithelial-to-mesenchymal transition enhances P-gp-mediated multidrug resistance in HCC827 cells. J Pharm Sci 106:2642–2649. https://doi.org/10.1016/j.xphs.2017.03.011
Tryndyak VP, Beland FA, Pogribny IP (2010) E-cadherin transcriptional down-regulation by epigenetic and microRNA-200 family alterations is related to mesenchymal and drug-resistant phenotypes in human breast cancer cells. Int J Cancer 126:2575–2583. https://doi.org/10.1002/ijc.24972
Vistain LF, Yamamoto N, Rathore R, Cha P, Meade TJ (2015) Targeted inhibition of snail activity in breast cancer cells by using a CoIII-Ebox conjugate. ChemBioChem 16:2065–2072. https://doi.org/10.1002/cbic.201500289
Wang C, Jin H, Wang N, Fan S, Wang Y, Zhang Y, Wei L, Tao X, Gu D, Zhao F, Fang J, Yao M, Qin W (2016) Gas6/Axl axis contributes to chemoresistance and metastasis in breast cancer through Akt/GSK-3β/β-catenin signaling. Theranostics 6:1205–1219. https://doi.org/10.7150/thno.15083
Wang X, Ling MT, Guan XY, Tsao SW, Cheung HW, Lee DT, Wong YC (2004) Identification of a novel function of Twist, a bHLH protein, in the development of acquired taxol resistance in human cancer cells. Oncogene 23:474–482. https://doi.org/10.1038/sj.onc.1207128
Wattanawongdon W, Hahnvajanawong C, Namwat N, Kanchanawat S, Boonmars T, Jearanaikoon P, Leelayuwat C, Techasen A, Seubwai W (2015) Establishment and characterization of gemcitabine-resistant human cholangiocarcinoma cell lines with multidrug resistance and enhanced invasiveness. Int J Oncol 47:398–410. https://doi.org/10.3892/ijo.2015.3019
Wilson C, Nicholes K, Bustos D, Lin E, Song Q, Stephan J-P, Kirkpatrick DS, Settleman J (2014) Overcoming EMT-associated resistance to anti-cancer drugs via Src/FAK pathway inhibition. Oncotarget 5:7328–7341. https://doi.org/10.18632/oncotarget.2397
Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M, Chan Z, Baron A, Franklin W, Drabkin HA, Girard L, Gazdar AF, Minna JD, Bunn PA (2006) Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res 66:944–950. https://doi.org/10.1158/0008-5472.CAN-05-1988
Wu Y, Wang Y, Lin Y, Liu Y, Wang Y, Jia J, Singh P, Chi Y-I, Wang C, Dong C, Li W, Tao M, Napier D, Shi Q, Deng J, Mark Evers B, Zhou BP (2017) Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting Snail1 degradation. Nat Commun 8:14228. https://doi.org/10.1038/ncomms14228
Xing H, Cao Y, Weng D, Tao W, Song X, Wang W, Meng L, Xu G, Zhou J, Wang S, Ma D (2008a) Fibronectin-mediated activation of Akt2 protects human ovarian and breast cancer cells from docetaxel-induced apoptosis via inhibition of the p38 pathway. Apoptosis 13:213–223. https://doi.org/10.1007/s10495-007-0158-5
Xing H, Weng D, Chen G, Tao W, Zhu T, Yang X, Meng L, Wang S, Lu Y, Ma D (2008b) Activation of fibronectin/PI-3K/Akt2 leads to chemoresistance to docetaxel by regulating survivin protein expression in ovarian and breast cancer cells. Cancer Lett 261:108–119. https://doi.org/10.1016/j.canlet.2007.11.022
Yada-Hashimoto N, Yamamoto T, Kamiura S, Seino H, Ohira H, Sawai K, Kimura T, Saji F (2003) Metastatic ovarian tumors: a review of 64 cases. Gynecol Oncol 89:314–317. https://doi.org/10.1016/S0090-8258(03)00075-1
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117:927–939. https://doi.org/10.1016/j.cell.2004.06.006
Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ (2008) Direct regulation of TWIST by HIF-1α promotes metastasis. Nat Cell Biol 10:295–305. https://doi.org/10.1038/ncb1691
Yeung TL, Leung CS, Yip KP, Yeung CLA, Wong STC, Mok SC (2015) Cellular and molecular processes in ovarian cancer metastasis. A review in the theme: cell and molecular processes in cancer metastasis. Am J Physiol 309:C444–C456. https://doi.org/10.1152/ajpcell.00188.2015
Yochum ZA, Cades J, Mazzacurati L, Neumann NM, Khetarpal SK, Chatterjee S, Wang H, Attar MA, Huang EHB, Chatley SN, Nugent K, Somasundaram A, Engh JA, Ewald AJ, Cho YJ, Rudin CM, Tran PT, Burns TF (2017) A first-in-class twist1 inhibitor with activity in oncogene-driven lung cancer. Mol Cancer Res 15:1764–1776. https://doi.org/10.1158/1541-7786.MCR-17-0298
Yochum ZA, Cades J, Wang H, Chatterjee S, Simons BW, O’Brien JP, Khetarpal SK, Lemtiri-Chlieh G, Myers KV, Huang EHB, Rudin CM, Tran PT, Burns TF (2019) Targeting the EMT transcription factor TWIST1 overcomes resistance to EGFR inhibitors in EGFR-mutant non-small-cell lung cancer. Oncogene 38:656–670. https://doi.org/10.1038/s41388-018-0482-y
Yoshida T, Song L, Bai Y, Kinose F, Li J, Ohaegbulam KC, Muñoz-Antonia T, Qu X, Eschrich S, Uramoto H, Tanaka F, Nasarre P, Gemmill RM, Roche J, Drabkin HA, Haura EB (2016) ZEB1 mediates acquired resistance to the epidermal growth factor receptor-tyrosine kinase inhibitors in non-small cell lung cancer. PLoS ONE 11:e0147344. https://doi.org/10.1371/journal.pone.0147344
Zhang F, Wang Z, Fan Y, Xu Q, Ji W, Tian R, Niu R (2015) Elevated STAT3 signaling-mediated upregulation of MMP-2/9 confers enhanced invasion ability in multidrug-resistant breast cancer cells. Int J Mol Sci 16:24772–24790. https://doi.org/10.3390/ijms161024772
Zhang F, Zhang H, Wang Z, Yu M, Tian R, Ji W, Yang Y, Niu R (2014) P-glycoprotein associates with Anxa2 and promotes invasion in multidrug resistant breast cancer cells. Biochem Pharmacol 87:292–302. https://doi.org/10.1016/j.bcp.2013.11.003
Zhang L, Li D, Yu S (2020) Pharmacological effects of harmine and its derivatives: a review. Arch Pharm Res 43:1259–1275. https://doi.org/10.1007/s12272-020-01283-6
Zhang W, Ding W, Chen Y, Feng M, Ouyang Y, Yu Y, He Z (2011) Up-regulation of breast cancer resistance protein plays a role in HER2-mediated chemoresistance through PI3K/Akt and nuclear factor-kappa B signaling pathways in MCF7 breast cancer cells. Acta Biochim Biophys Sin 43:647–653. https://doi.org/10.1093/abbs/gmr050
Zhang W, Feng M, Zheng G, Chen Y, Wang X, Pen B, Yin J, Yu Y, He Z (2012) Chemoresistance to 5-fluorouracil induces epithelial-mesenchymal transition via up-regulation of Snail in MCF7 human breast cancer cells. Biochem Biophys Res Commun 417:679–685. https://doi.org/10.1016/j.bbrc.2011.11.142
Zhang X, Zhang Z, Zhang Q, Zhang Q, Sun P, Xiang R, Ren G, Yang S (2018) ZEB1 confers chemotherapeutic resistance to breast cancer by activating ATM. Cell Death Dis 9:57. https://doi.org/10.1038/s41419-017-0087-3
Zhao X, Wang D, Ding Y, Zhou J, Liu G, Ji Z (2019) LncRNA ZEB1-AS1 promotes migration and metastasis of bladder cancer cells by post-transcriptional activation of ZEB1. Int J Mol Med 44:196–206. https://doi.org/10.3892/ijmm.2019.4187
Zhu C, Wei Y, Wei X (2019) AXL receptor tyrosine kinase as a promising anti-cancer approach: functions, molecular mechanisms and clinical applications. Mol Cancer 18:153. https://doi.org/10.1186/s12943-019-1090-3
Zhu X, Shen H, Yin X, Long L, Xie C, Liu Y, Hui L, Lin X, Fang Y, Cao Y, Xu Y, Li M, Xu W, Li Y (2016) MiR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene 35:323–332. https://doi.org/10.1038/onc.2015.84
Zhuo WL, Wang Y, Zhuo XL, Zhang YS, Chen ZT (2008) Short interfering RNA directed against TWIST, a novel zinc finger transcription factor, increases A549 cell sensitivity to cisplatin via MAPK/mitochondrial pathway. Biochem Biophys Res Commun 369:1098–1102. https://doi.org/10.1016/j.bbrc.2008.02.143
Acknowledgements
This research was supported by National Research Foundation of Korea (NRF) grants, funded by the Korea government (MSIT) (2021R1A2C1011196 and 2015R1A5A1008958). This research was supported by the Chung-Ang University Graduate Research Scholarship in 2018.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Seo, J., Ha, J., Kang, E. et al. The role of epithelial–mesenchymal transition-regulating transcription factors in anti-cancer drug resistance. Arch. Pharm. Res. 44, 281–292 (2021). https://doi.org/10.1007/s12272-021-01321-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-021-01321-x