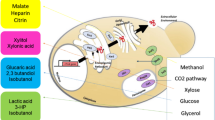
Abstract
Cell-free systems utilize a subset of cellular components without intact cell wall and/or membranes. The system was first invented for use in fermentation. Subsequent improvements enabled its application in protein synthesis, which is still the most common use of the system. Lately, attempts have been reported where metabolic engineering concepts and techniques were applied to cell-free systems and/or vice-versa. These attempts and advances led to exciting discoveries about biochemical reactions, as well as properties and/or structures of cellular components that make up complex biological systems. This review will first provide a basic overview and brief history of the cell-free system. Then, explanation on recent advances in the field will be provided, followed by notes on the innovative applications. Future outlook of the field will also be covered with the emphasis on how the emerging data science methods can be applied to improve the system and its applicability.
Similar content being viewed by others
References
Swartz, J. R. (2018) Expanding biological applications using cell-free metabolic engineering: An overview. Metab. Eng. 50: 156–172.
Buchner, E. (1897) Alkoholische Gährung ohne Hefezellen. Ber. Dtsch. Chem. Ges. 30: 117–124.
Nirenberg, M. W. and H. J. Matthaei (1961) The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Nat. Acad. Sci. USA. 47: 1588–1602.
Bechtold, M., E. Brenna, C. Femmer, F. G. Gatti, S. Panke, F. Parmeggiani, and A. Sacchetti (2012) Biotechnological development of a practical synthesis of ethyl (S)-2-ethoxy-3-(p-methoxyphenyl)propanoate (EEHP): Over 100-fold productivity increase from yeast whole cells to recombinant isolated enzymes. Org. Process Res. Dev. 16: 269–276.
Korman, T. P., P. H. Opgenorth, and J. U. Bowie (2017) A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nat. Comm. 8: 15526.
Takors, R. (2012) Scale-up of microbial processes: Impacts, tools and open questions. J. Biotechnol. 160: 3–9.
Calhoun, K. and J. R. Swartz (2006) Total amino acid stabilization during cell-free protein synthesis reactions. J. Biotechnol. 123: 193–203.
Shrestha, P., T. M. Holland, and B. C. Bundy (2012) Streamlined extract preparation for Escherichia coli-based cell-free protein synthesis by sonication or bead vortex mixing. Biotechniques. 53: 163–174.
Didovyk, A., T. Tonooka, L. Tsimring, and J. Hasty (2017) Rapid and scalable preparation of bacterial lysates for cell-free gene expression. ACS Synth. Biol. 6: 2198–2208.
Kim, D. M. and J. R. Swartz (2000) Oxalate improves protein synthesis by enhancing ATP supply in a cell-free system derived from Escherichia coli. Biotechnol. Lett. 22: 1537–1542.
Nagumo, Y., K. Fujiwara, K. Horisawa, H. Yanagawa, and N. Doi (2016) PURE mRNA display for in vitro selection of single-chain antibodies. J. Biochem. 159: 519–526.
Smith, M. T., S. D. Berkheimer, C. J. Werner, and B. C. Bundy (2014) Lyophilized Escherichia coli-based cell-free systems for robust, high-density, long-term storage. Biotechniques. 56: 186–193.
Liu, D. V., J. F. Zawada, and J. R. Swartz (2005) Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis. Biotechnol. Prog. 21: 460–465.
Lu, Y. (2017) Cell-free synthetic biology: Engineering in an open world. Synth. Sys. Bioltechnol. 2: 23–27.
Lim, H. J. and D. M. Kim (2019) Cell-free metabolic engineering: Recent developments and future prospects. Methods Protoc. 2: 33.
Brodel, A. K., A. Sonnabend, and S. Kubick (2014) Cell-free protein expression based on extracts from CHO cells. Biotechnol. Bioeng. 111: 25–36.
Ezure, T., K. Nanatani, Y. Sato, S. Suzuki, K. Aizawa, S. Souma, M. Ito, T. Hohsaka, G. von Heijine, T. Utsumi, K. Abe, E. Ando, and N. Uozumi (2014) A cell-free translocation system using extracts of cultured insect cells to yield functional membrane proteins. PLoS One. 9: e112874.
Takai, K., T. Sawasaki, and Y. Endo (2010) Practical cell-free protein synthesis system using purified wheat embryos. Nat. Protoc. 5: 227–238.
Kuruma, Y. and T. Ueda (2015) The PURE system for the cell-free synthesis of membrane proteins. Nat. Protoc. 10: 1328–1344.
Michel-Reydellet, N., K. Calhoun, and J. R. Swartz (2004) Amino acid stabilization for cell-free protein synthesis by modification of the Escherichia coli genome. Metab. Eng. 6: 197–203.
Giel, J. L., D. Rodionov, M. Liu, F. R. Blattner, and P. J. Kiley (2006) IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O-regulated genes in Escherichia coli. Mol. Microbiol. 60: 1058–1075.
Boyer, M. E., J. A. Stapleton, J. M. Kuchenreuther, C. W. Wang, and J. R. Swartz (2008) Cell-free synthesis and maturation of [FeFe] hydrogenases. Biotechnol. Bioeng. 99: 59–67.
De La Paz, L. (2016) Elucidating in vitro Activation of [FeFe] Hydrogenase. Ph.D. Thesis. Stanford University, Stanford, CA, USA.
Peters, J. W., W. N. Lanzilotta, B. J. Lemon, and L. C. Seefeldt (1998) X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 282: 1853–1858.
Kuchenreuther, J. M., W. K. Myers, T. A. Stich, S. J. George, Y. Nejatyjahromy, J. R. Swartz, and R. D. Britt (2013) A radical intermediate in tyrosine scission to the CO and CN- ligands of FeFe hydrogenase. Science. 342: 472–475.
Kuchenreuther, J. M., W. K. Myers, D. L. M. Suess, T. A. Stich, V. Pelmenschikov, S. A. Shiigi, S. P. Cramer, J. R. Swartz, R. D. Britt, and S. J. George (2014) The HydG enzyme generates an Fe(CO)2(CN) synthon in assembly of the FeFe hydrogenase H-cluster. Science. 343: 424–427.
Dinis, P., D. L. M. Suess, S. J. Fox, J. E. Harmer, R. C. Driesener, L. De La Paz, J. R. Swartz, J. W. Essex, R. D. Britt, and P. L. Roach (2015) X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly. Proc. Nat. Acad. Sci. USA. 112: 1362–1367.
Lu, Y., J. P. Welsh, W. Chan, and J. R. Swartz (2013) Escherichia coli-based cell free production of flagellin and ordered flagellin display on virus-like particles. Biotechnol. Bioeng. 110: 2073–2085.
Wuu, J. J. and J. R. Swartz (2008) High yield cell-free production of integral membrane proteins without refolding or detergents. Biochim. Biophys. Acta. 1778: 1237–1250.
Schoborg, J. A., J. M. Hershewe, J. C. Stark, W. Kightlinger, J. E. Kath, T. Jaroentomeechai, A. Natarajan, M. P. DeLisa, and M. C. Jewett (2018) A cell-free platform for rapid synthesis and testing of active oligosaccharyltransferases. Biotechnol. Bioeng. 115: 739–750.
Kolb, V. A., E. V. Makeyev, W. W. Ward, and A. S. Spirin (1996) Synthesis and maturation of green fluorescent protein in a cell-free translation system. Biotechnol. Lett. 18: 1447–1452.
Kuchenreuther, J. M., S. A. Shiigi, and J. R. Swartz (2014) Cellfree synthesis of the H-Cluster: A model for the in vitro assembly of metalloprotein metal centers. pp. 49–72. In: J. C. Fontecilla-Camps and Y. Nicolet (eds.). Metalloproteins. Humana Press, Totowa, NJ, USA.
Oza, J. P., H. R. Aerni, N. L. Pirman, K. W. Barber, C. M. Ter Haar, S. Rogulina, M. B. Amrofell, F. J. Isaacs, J. Rinehart, and M. C. Jewett (2015) Robust production of recombinant phosphoproteins using cell-free protein synthesis. Nat. Commun. 6: 8168.
Yang, J. P., T. Cirico, F. Katzen, T. C. Peterson, and W. Kudlicki (2011) Cell-free synthesis of a functional G protein-coupled receptor complexed with nanometer scale bilayer discs. BMC Biotechnol. 11: 57.
Young, E. T. (1970) Cell-free synthesis of bacteriophage T4 glucosyl transferase. J. Mol. Biol. 51: 591–604.
Min, S. E., K. H. Lee, S. W. Park, T. H. Yoo, C. H. Oh, J. H. Park, S. Y. Yang, Y. S. Kim, and D. M. Kim (2016) Cell-free production and streamlined assay of cytosol-penetrating antibodies. Biotechnol. Bioeng. 113: 2107–2112.
Martin, R. W., N. I. Majewska, C. X. Chen, T. E. Albanetti, R. B. C. Jimenez, A. E. Schmelzer, M. C. Jewett, and V. Roy (2017) Development of a CHO-based cell-free platform for synthesis of active monoclonal antibodies. ACS Synth. Biol. 6: 1370–1379.
Stech, M., O. Nikolaeva, L. Thoring, W. F. M. Stöcklein, D. A. Wüstenhagen, M. Hust, S. Dübel, and S. Kubick (2017) Cell-free synthesis of functional antibodies using a coupled in vitro transcription-translation system based on CHO cell lysates. Sci. Rep. 7: 12030.
Takeda, H., T. Ogasawara, T. Ozawa, A. Muraguchi, P. J. Jih, R. Morishita, M. Uchigashima, M. Watanabe, T. Fujimoto, T. Iwasaki, Y. Endo, and T. Sawasaki (2015) Production of monoclonal antibodies against GPCR using cell-free synthesized GPCR antigen and biotinylated liposome-based interaction assay. Sci. Rep. 5: 11333.
Koo, J., S. Shiigi, M. Rohovie, K. Mehta, and J. R. Swartz (2016) Characterization of [FeFe] hydrogenase O sensitivity using a new, physiological approach. J. Biol. Chem. 291: 21563–21570.
Koo, J., T. Schnabel, S. Liong, N. H. Evitt, and J. R. Swartz (2017) High-throughput screening of catalytic H production. Angew. Chem. Int. Ed. Engl. 56: 1012–1016.
Koo, J. and J. R. Swartz (2018) System analysis and improved [FeFe] hydrogenase O tolerance suggest feasibility for photosynthetic H production. Metab. Eng. 49: 21–27.
Liu, C. C. and P. G. Schultz (2010) Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79: 413–444.
Moses, J. E. and A. D. Moorhouse (2007) The growing applications of click chemistry. Chem. Soc. Rev. 36: 1249–1262.
Kolb, H. C., M. G. Finn, and K. G Sharpless (2001) Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 40: 2004–2021.
Baskin, J. M., J. A. Prescher, S. T. Laughlin, N. J. Agard, P. V. Chang, I. A. Miller, A. Lo, J. A. Codelli, and C. R. Bertozzi (2007) Copper-free click chemistry for dynamic in vivo imaging. Proc. Nat. Acad. Sci. USA. 104: 16793–16797.
Wang, F., W. Niu, J. Guo, and P. G. Schultz (2012) Unnatural amino acid mutagenesis of fluorescent proteins. Angew. Chem. Int. Ed. Engl. 51: 10132–10135.
Wong, H. E., S. P. Pack, and I. Kwon (2016) Positional effects of hydrophobic non-natural amino acid mutagenesis into the surface region of murine dihydrofolate reductase on enzyme properties. Biochem. Eng. J. 109: 1–8.
Key, H. M., P. Dydio, D. S. Clark, and J. F. Hartwig (2016) Abiological catalysis by artificial haem proteins containing noble metals in place of iron. Nature. 534: 534–537.
Dydio, P., H. M. Key, A. Nazarenko, J. Y. E. Rha, V. Seyedkazemi, D. S. Clark, and J. F. Hartwig (2016) An artificial metalloenzyme with the kinetics of native enzymes. Science. 354: 102–106.
Bailey, J. B., R. H. Subramanian, L. A. Churchfield, and F. A. Tezcan (2016) Metal-directed design of supramolecular protein assemblies. Methods Enzymol. 580: 223–250.
Zhang, P., H. Feng, J. Yang, H. Jiang, H. Zhou, and Y. Lu (2019) Detection of inorganic ions and organic molecules with cell-free biosensing systems. J. Biotechnol. 300: 78–86.
Jang, Y. J., K. H. Lee, T. H. Yoo, and D. M. Kim (2019) Interfacing a personal glucose meter with cell-free protein synthesis for rapid analysis of amino acids. Anal. Chem. 91: 2531–2535.
Byun, J. Y., K. H. Lee, Y. B. Shin, and D. M. Kim (2019) Cascading amplification of immunoassay signal by cell-free expression of firefly luciferase from detection antibody-conjugated DNA in an Escherichia coli extract. ACS Sens. 4: 93–99.
Pardee, K., S. Slomovic, P. Q. Nguyen, J. W. Lee, N. Donghia, D. Burrill, T. Ferrante, F. R. McSorley, Y. Furuta, A. Vernet, M. Lewandowski, C. N. Boddy, N. S. Joshi, and J. J. Collins (2016) Portable, on-demand biomolecular manufacturing. Cell. 167: 248–259.e12.
Luna, J. M., R. D. Rufino, and L. A. Sarubbo (2016) Biosurfactant from Candida sphaerica UCP0995 exhibiting heavy metal remediation properties. Process Saf. Environ. Prot. 102: 558–566.
Park, Y. J., H. J. Choi, and W. H. Joo (2017) Effects of cell-free solutions of probiotic bacteria on putrescine production by food borne-pathogens in ornithine decarboxylase broth. Proceedings of the 2017 KSBB Fall Meeting and International Symposium. October 11–13. Busan, Korea.
Kightlinger, W., K. E. Duncker, A. Ramesh, A. H. Thames, A. Natarajan, J. C. Stark, A. Yang, L. Lin, M. Mrksich, M. P. DeLisa, and M. C. Jewett (2019) A cell-free biosynthesis platform for modular construction of protein glycosylation pathways. Nat. Commun. 10: 5404.
Jaroentomeechai, T., J. C. Stark, A. Natarajan, C. J. Glasscock, L. E. Yates, K. J. Hsu, M. Mrksich, M. C. Jewett, and M. P. DeLisa (2018) Single-pot glycoprotein biosynthesis using a cell-free transcription-translation system enriched with glycosylation machinery. Nat. Commun. 9: 2686.
Karim, A. S. and M. C. Jewett (2018) Cell-free synthetic biology for pathway prototyping. Methods Enzymol. 608: 31–57.
Acknowledgements
This study was supported by the Hongik University new faculty research support fund.
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Koo, J., Yang, J. & Park, H. Cell-free Systems: Recent Advances and Future Outlook. Biotechnol Bioproc E 25, 955–961 (2020). https://doi.org/10.1007/s12257-020-0013-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-020-0013-x