Abstract
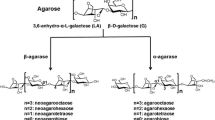
An agar-degrading bacterium, strain S1, was isolated from the coastal seawater of Jeju Island, Korea, and identified as a novel species of the genus Vibrio. The isolate, Vibrio sp. S1, produced at least five kinds of extracellular agarases in artificial sea water broth containing yeast extract and bacto peptone, and two of them were purified to homogeneity. Both agarases, AgaA33 and AgaA29, with apparent molecular weights of 33 kDa and 29 kDa, respectively, exhibited an optimum temperature and pH of 45°C and 7.0, respectively. AgaA33 and AgaA29 showed acidophilic properties and maintained 93% and 87% of the maximum agarase activity at 50°C, respectively, displaying their thermostability. Moreover, more than 80% activity was retained after heat treatment at 45°C for 1 h. Their agarase activities were inhibited by the presence of EDTA and remarkably stimulated by the presence of Mn2+ in a concentration-dependent manner, indicating that both agarases required the Mn2+ ion as a cofactor. The AgaA33 enzyme exhibited Km and Vmax values of 4.02 mg/mL and 27 U/mg, respectively. AgaA29 exhibited Km and Vmax values of 3.26 mg/mL and 200 U/mg, respectively. The instrumental analysis demonstrated that both are new β-agarases that can hydrolyze agarose and agaro-oligomers into neoagarotetraose and neoagarohexaose. In addition, AgaA33 coproduced neoagarooctaose as the major final product. Both thermostable enzymes are expected to be useful for the industrial application of agar.
Similar content being viewed by others
References
Rhein-Knudsen, N., M. T. Ale, and A. S. Meyer (2015) Seaweed hydrocolloid production: an update on enzyme assisted extraction and modification technologies. Mar. Drugs. 13: 3340–3359.
Chi, W. J., Y. K. Chang, and S. K. Hong (2012) Agar degradation by microorganisms and agar-degrading enzymes. Appl. Microbiol. Biotechnol. 94: 917–930.
Usov, A. I. (2011) Polysaccharides of the red algae. Adv. Carbohydr. Chem. Biochem. 65: 115–217.
Nussinovitch, A. (1997) Hydrocolloid Applications: Gum technology in the food and other industries. pp. 1–18. Springer, New York, NY, USA.
Brinker, C. J. and G. W. Scherer (2013) Sol-gel science: the physics and chemistry of sol-gel processing. pp. 2–10. Sol-gel processing. Academic Press, Cambridge, MA, USA.
Kim, J. H., E. J. Yun, N. Seo, S. Yu, D. H. Kim, K. M. Cho, H. J. An, J. H. Kim, I. G. Choi, and K. H. Kim (2017) Enzymatic liquefaction of agarose above the sol-gel transition temperature using a thermostable endo-type β-agarase, Aga16B. Appl. Microbiol. Biotechnol. 101: 1111–1120.
Kim, H. T., S. Lee, K. H. Kim, and I. G. Choi (2012) The complete enzymatic saccharification of agarose and its application to simultaneous saccharification and fermentation of agarose for ethanol production. Bioresour. Technol. 107: 301–306.
Kim, D. H., E. J. Yun, S. H. Lee, and K. H. Kim (2018) Novel two-step process utilizing a single enzyme for the production of high-titer 3,6-anhydro-L-galactose from agarose derived from red macroalgae. J. Agric. Food. Chem. 66: 12249–12256.
Park, D. Y., W. J. Chi, J. S. Park, Y. K. Chang, and S. K. Hong (2015) Cloning, expression, and biochemical characterization of a GH16 β-agarase AgaH71 from Pseudoalteromonas hodoensis H7. Appl. Biochem. Biotechnol. 175: 733–747.
Baker, G. C., J. J. Smith, and D. A. Cowan (2003) Review and reanalysis of domain-specific 16S primers. J. Microbiol. Methods. 55: 541–555.
Chun, J., J. H. Lee, Y. Jung, M. Kim, S. Kim, B. K. Kim, and Y. W. Lim (2007) ExTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57: 2259–2261.
Thomson, J. D., D. G. Higgins, and T. J. Gibson (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680.
Hall, T. A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98.
Saitou, N. and M. Nei (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425.
Tamura, K., G. Stecher, D. Peterson, A. Filipski, and S. Kumar (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729.
Kimura, M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120.
Komagata, K. and K. Suzuki (1987) Lipid and cell-wall analysis in bacterial systematic. Methods Microbiol. 19: 161–207.
Mesbah, M., U. Premachandran, and W. B. Whitman (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39: 159–167.
Sasser, M. (1997) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark, DE, USA.
Lineweaver, H. and D. Burk (1934) The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56: 658–666.
Temuujin, U., W. J. Chi, S. Y. Lee, Y. K. Chang, and S. K. Hong (2011) Overexpression and biochemical characterization of DagA from Streptomyces coelicolor A3(2): an endo-type β-agarase producing neoagarotetraose and neoagarohexaose. Appl. Microbiol. Biotechnol. 92: 749–759.
Stackegrandt, E. and J. Ebers (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today. 33: 152–155.
Morrice, L. M., M. W. McLean, F. B. Williamson, and W. F. Long (1983) Beta agarase I and II from Pseudomonas atlantica purifications and some properties. Eur. J. Biochem. 135: 553–558.
Rochas, C., P. Potin, and B. Kloareg (1994) NMR spectroscopic investigation of agarose oligomers produced by an alpha-agarase. Carbohydr. Res. 253: 69–77.
Hong, S. J., J. H. Lee, E. J. Kim, H. J. Yang, J. S. Park, and S. K. Hong (2017) Anti-obesity and anti-diabetic effect of neoagarooligosaccharides on high-fat diet-induced obesity in mice. Mar. Drugs. 15: 90.
Li, J., Y. Sha, D. Seswita-Zilda, Q. Hu, and P. He (2014) Purification and characterization of thermostable agarase from Bacillus sp. BI-3, a thermophilic bacterium isolated from hot spring. J. Microbiol. Biotechnol. 24: 19–25.
Bannikova, G. E., S. A. Lopatin, V. P. Varlamov, B. B. Kuznetsov, I. V. Kozina, M. L. Miroshnichenko, N. A. Chernykh, T. P. Turova, and E. A. Bonch-Osmolovskaia. (2008) The thermophilic bacteria hydrolyzing agar: characterization of thermostable agarase. Prikl. Biokhim. Mikrobiol. 44: 404–409.
Minegishi, H., Y. Shimane, A. Echigo, Y. Ohta, Y. Hatada, M. Kamekura, T. Maruyama, and R. Usami (2013) Thermophilic and halophilic β-agarase from a halophilic archaeon Halococcus sp. 197A. Extremophiles. 17: 931–939.
Di, W., W. Qu, and R. Zeng (2018) Cloning, expression, and characterization of thermal-stable and pH-stable agarase from mangrove sediments. J. Basic. Microbiol. 58: 302–309.
Chen, Z. W., H. J. Lin, W. C. Huang, S. L. Hsuan, J. H. Lin, and J. P. Wang (2018) Molecular cloning, expression, and functional characterization of the β-agarase AgaB-4 from Paenibacillus agarexedens. AMB Express. 8: 49.
Chi, W. J., C. R. Lee, S. Dugerjonjuu, J. S. Park, D. K. Kang, and S. K. Hong (2015) Biochemical characterization of a novel iron-dependent GH16 β-agarase, AgaH92, from an agarolytic bacterium Pseudoalteromonas sp. H9. FEMS Microbiol. Lett. 362: fnv035.
Ramos, K. R. M., K. N. G. Valdehuesa, G. M. Nisola, W. K. Lee, and W. J. Chung (2018) Identification and characterization of a thermostable endolytic β-agarase Aga2 from a newly isolated marine agarolytic bacteria Cellulophaga omnivescoria W5C. N. Biotechnol. 40: 261–267.
Yang, J. H., S. S. Cho, K. M. Kim, J. Y. Kim, E. J. Kim, E. Y. Park, J. H. Lee, and S. H. Ki (2017) Neoagarooligosaccharides enhance the level and efficiency of LDL receptor and improve cholesterol homeostasis. J. Funct. Foods. 38: 529–539.
Kang, D. R., G. Y. Yoon, J. Cho, S. J. Lee, S. J. Lee, H. J. Park, T. H. Kang, H. D. Han, W. S. Park, Y. K. Yoon, Y. M. Park, and I. D. Jung (2017) Neoagarooligosaccharides prevent septic shock by modulating A20- and cyclooxygenase-2-mediated interleukin-10 secretion in a septic-shock mouse model. Biochem. Biophys. Res. Commun. 486: 998–1004.
Lee, M. H., J. H. Jang, G. Y. Yoon, S. J. Lee, M. G. Lee, T. H. Kang, H. D. Han, H. S. Kim, W. S. Choi, W. S. Park, Y. M. Park, and I. D. Jung (2017) Neoagarohexaose-mediated activation of dendritic cells via Toll-like receptor 4 leads to stimulation of natural killer cells and enhancement of antitumor immunity. BMB Rep. 50: 263–268.
Zhang, N., X. Mao, R. W. Li, E. Hou, Y. Wang, C. Xue, and Q. Tang (2017) Neoagarotetraose protects mice against intense exercise-induced fatigue damage by modulating gut microbial composition and function. Mol. Nutr. Food Res. 61: 1600585.
Lee, J. S., S. K. Hong, C. R. Lee, S. W. Nam, S. J. Jeon, and Y. H. Kim (2019) Production of ethanol (agaro-bioethanol) from agarose by unified enzymatic saccharification and fermentation in recombinant yeast. J. Microbiol. Biotechnol. 29: 625–632.
Aoki, T., T. Araki, and M. Kitamikado (1990) Purification and characterization of a novel beta-agarase from Vibrio sp. AP-2. Eur. J. Biochem. 187: 461–465.
Sugano, Y., I. Terada, M. Arita, M. Noma, and T. Matsumoto (1993) Purification and characterization of a new agarase from a marine bacterium, Vibrio sp. Strain JT0107. Appl. Environ. Microbiol. 59: 1549–1554.
Dong, J., Y. Tamaru, and T. Araki (2007) A unique beta-agarase, AgaA, from a marine bacterium, Vibrio sp. strain PO-303. Appl. Microbiol. Biotechnol. 74: 1248–55.
Liao, L., X. W. Xu, X. W. Jiang, Y. Cao, N. Yi, Y. Y. Huo, Y. H. Wu, X. F. Zhu, X. Q. Zhang, and M. Wu (2011) Cloning, expression, and characterization of a new beta-agarase from Vibrio sp. strain CN41. Appl. Environ. Microbiol. 77: 7077–7079.
Acknowledgement
This work was supported by the Advanced Biomass R and D Center (ABC) of Global Frontier Project funded by the Ministry of Science, ICT and Future Planning (NRF-2015M3A6A2065700).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chi, WJ., Seo, J.W. & Hong, SK. Characterization of Two Thermostable β-agarases from a Newly Isolated Marine Agarolytic Bacterium, Vibrio sp. S1. Biotechnol Bioproc E 24, 799–809 (2019). https://doi.org/10.1007/s12257-019-0180-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-019-0180-9