Abstract
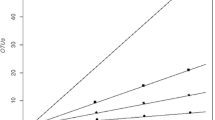
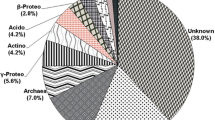
The objectives of this study were (i) to isolate and characterize of cultivable denitrifying bacteria using classic microbiological and molecular methods, (ii) to compare of 16S rRNA and nosZ genes as molecular markers, (iii) to determine bacterial community structure and diversity in soil samples using single-strand conformation polymorphism (SSCP) analysis. In this study, 49 bacterial isolates were cultivated and phylogenetic analyses grouped them into two phyla: Proteobacteria (37 species) and Firmicutes (12 species). Our study showed that the nosZ functional gen could be used to identify denitrifying bacteria abundance in environment but could not be used to identify pure bacterial cultures. In addition, the bacterial community structure showed significant differences among the various soil types. Phylogenetic analysis of community structure indicated that 51 clones could be divided into 2 phylotypes. Uncultured bacteria (80.4%) and Gammaproteobacteria (19.6%) were the dominant components of the soil bacterial community. For 16S rRNA, PCR products of 49 bacteria were obtained with 27F-1492R primer pairs. For nosZ, PCR products were obtained with primers 1F-1R (259 bp), 2F-2R (267 bp), and F-1622R (453 bp) of 39 bacteria that the single nosZ band provided on the agarose gel. The bacterial 16S rRNA gene clone library was dominated by Gammaproteobacteria and Bacilli. The nosZ clone sequences did not represent the bacteria from which they were obtained but were found to be closer to the environmental clones. Our study showed that the nosZ functional gene could be used to identify denitrification abundance in environment but could not be used to identify pure bacterial cultures. It was also found that the nosZ sequences showed uncultured denitrifier species.





Similar content being viewed by others
References
Avşar C (2018) Quantification of denitrifier population size in soil, bacteria community structures and comparison of nosZ and 16S rRNA genes from culturable denitrifying. PhD Thesis, Ankara University, Ankara, Turkey, 188p.
Awong-Taylor J, Craven KS, Griffiths L, Bass C, Muscarella M (2008) Comparison of biochemical and molecular methods for the identification of bacterial isolates associated with failed loggerhead sea turtle eggs. J Appl Microbiol 104(5):1244–1251. https://doi.org/10.1111/j.1365-2672.2007.03650.x
Backman JS, Hermansson A, Tebbe CC, Lindgren PE (2003) Liming induces growth of a diverse flora of ammonia-oxidising bacteria in acid spruce forest soil as determined by SSCP and DGGE. Soil Bio Biochem 35(10):1337–1347. https://doi.org/10.1016/s0038-0717(03)00213-x
Belak Á, Kovács M, Hermann Z, Holczman Á, Márta D, Stojakovič S, Maraz A (2011) Molecular analysis of poultry meat spoiling microbiota and heterogeneity of their proteolytic and lipolytic enzyme activities. Acta Aliment 40(1):3–22. https://doi.org/10.1556/aalim.40.2011.suppl.2
Benga L, Benten WPM, Engelhardt E, Köhrer K, Gougoula C, Sager M (2014) 16S ribosomal DNA sequence-based identification of bacteria in laboratory rodents: a practical approach in laboratory animal bacteriology diagnostics. Lab Anim 48(4):305–312. https://doi.org/10.1177/0023677214538240
Bosshard PP, Abels S, Altwegg M, Böttger EC, Zbinden R (2004) Comparison of conventional and molecular methods for identification of aerobic catalase-negative Gram-positive cocci in the clinical laboratory. J Clin Microbiol 42(5):2065–2073. https://doi.org/10.1128/jcm.42.5.2065-2073.2004
Bowles MW, Nigro LM, Teske AP, Joye SB (2012) Denitrification and environmental factors influencing nitrate removal in Guaymas Basin hydrothermally altered sediments. Front Microbiol 3:1–11. https://doi.org/10.3389/fmicb.2012.00377
Bunge M, Lechner U (2004) Studying the microbial dynamics of a trichlorobenzene-dechlorinating community by single-strand conformation polymorphism. Kapitel 6:127–140. https://doi.org/10.1002/0471684228.egp11609
Byun SO, Fang Q, Zhou H, Hickford JGH (2009) An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Analytic Biochem 385(1):174–175. https://doi.org/10.1016/j.ab.2008.10.024
Carlson JM, Leonard AB, Hyde ER, Petrosino JF, Primm TP (2017) Microbiome disruption and recovery in the fish Gambusia affinis following exposure to broad-spectrum antibiotic. Infect Drug Res 10:143. https://doi.org/10.2147/idr.s129055
Čuhel J, Šimek M, Laughlin RJ, Bru D, Chèneby D, Watson CJ, Philippot L (2010) Insights into the effect of soil pH on N2O and N2 emissions and denitrifier community size and activity. Appl Environ Microbiol 76(6):1870–1878. https://doi.org/10.1128/aem.02484-09
Cui P, Fan F, Yin C, Song A, Huang P, Tang Y, Liang Y (2016) Long-term organic and inorganic fertilization alters temperature sensitivity of potential N2O emissions and associated microbes. Soil Bio Biochem 93:131–141. https://doi.org/10.1016/j.soilbio.2015.11.005
Dandie CE, Burton DL, Zebarth BJ, Trevors JT, Goyer C (2007) Analysis of denitrification genes and comparison of nosZ, cnorB, and 16S rDNA from culturable denitrifying bacteria in potato cropping systems. Syst Appl Microbiol 30:128–138. https://doi.org/10.1016/j.syapm.2006.05.002
Delorme S, Philippot L, Edel-Hermann V, Deulvot C, Mougel C, Lemanceau P (2003) Comparative genetic diversity of the narG, nosZ, and 16S rRNA genes in fluorescent pseudomonads. Appl Environ Microbiol 69(2):1004–1012. https://doi.org/10.1128/aem.69.2.1004-1012.2003
Gaimster H, Alston M, Richardson D, Gates A, Rowley G (2017) Transcriptional and environmental control of bacterial denitrification and N2O emissions. FEMS Microbiol Let fnx277. https://doi.org/10.1093/femsle/fnx277
Graf DRH, Jones CM, Hallin S (2014) Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS ONE 9:e114118. https://doi.org/10.1371/journal.pone.0114118
Hayatsu M, Tago K, Saito M (2008) Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci Plant Nut 54:33–45. https://doi.org/10.1111/j.1747-0765.2007.00195.x
Henry S, Bru D, Stres B, Hallet S, Philippot L (2006) Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soil. Appl Environ Microbiol 72:5181–5189. https://doi.org/10.1128/aem.00231-06
Heylen K, Gevers D, Vanparys B, Wittebolle L, Geets J, Boon N (2006) The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ Microbiol 8:2012–2021. https://doi.org/10.1111/j.1462-2920.2006.01081.x
Hori T, Haruta S, Ueno Y, Ishii M, Igarashi Y (2006) Direct comparison of single-strand conformation polymorphism (SSCP) and denaturing gradient gel electrophoresis (DGGE) to characterize a microbial community on the basis of 16S rRNA gene fragments. J Microbiol Meth 66:165–169. https://doi.org/10.1016/j.mimet.2005.11.007
Horn MA, Drake HL, Schramm A (2006) Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar. Appl Environ Microbiol 72(2):1019–1026. https://doi.org/10.1128/aem.72.2.1019-1026.2006
Ishii S, Ashida N, Otsuka S, Senoo K (2011) Isolation of oligotrophic denitrifiers carrying previously uncharacterized functional gene sequences. Appl Environ Microbiol 77(1):338–342. https://doi.org/10.1128/aem.02189-10
Jones CM, Graf DRH, Bru D, Philippot L, Hallin S (2013) The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. The ISME J 7:417–426. https://doi.org/10.1038/ismej.2012.125
Mills HJ, Hunter E, Humphrys M, Kerkhof L, McGuinness L, Huettel M, Kostka JE (2008) Characterization of nitrifying, denitrifying, and overall bacterial communities in permeable marine sediments of the northeastern Gulf of Mexico. Appl Environ Microbiol 74(14):4440–4453. https://doi.org/10.1128/aem.02692-07
Morkved PT, Dörsch P, Bakken LR (2007) The N2O product ratio of nitrification and its dependence on long-term changes in soil pH. Soil Bio Biochemist 39(8):2048–2057. https://doi.org/10.1016/j.soilbio.2007.03.006
Morley N, Baggs EM, Dörsch P, Bakken L (2008) Production of NO, N2O and N2 by extracted soil bacteria, regulation by NO2− and O2 concentrations. FEMS Microbe Eco 65(1):102–112. https://doi.org/10.1111/j.1574-6941.2008.00495.x
Philippot L, Hallin S, Schloter M (2007) Ecology of denitrifying prokaryotes in agricultural soil. Adv Agro 96:249–305. https://doi.org/10.1016/s0065-2113(07)96003-4
Philippot L, Spor A, Henault C, Bru D, Bizouard F, Jones CM, Sarr A, Maron P (2013) Loss in microbial diversity affects nitrogen cycling in soil. The ISME J 7:1609–1619. https://doi.org/10.1038/ismej.2013.34
Pihlatie M, Syväsalo E, Simojoki A, Esala M, Regina K (2004) Contribution of nitrification and denitrification to N2O production in peat, clay and loamy sand soils under different soil moisture conditions. Nut Cycle Agro 70(2):135–141. https://doi.org/10.1023/b:fres.0000048475.81211.3c
Richardson D, Felgate H, Watmough N, Thomson A, Baggs E (2009) Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle could enzymic regulation hold the key. Trends Biotech 27:388–397. https://doi.org/10.1016/j.tibtech.2009.03.009
Rossmann B, Müller H, Smalla K, Mpiira S, Tumuhairwe JB, Staver C, Berg G (2012) Banana-associated microbial communities in Uganda are highly diverse but dominated by Enterobacteriaceae. Appl Environ Microbiol 78(14):4933–4941. https://doi.org/10.1128/aem.00772-12
Saggar S, Jha N, Deslippe J, Bolan NS, Luo J, Giltrap DL, Tillman RW (2013) Denitrification and N2O:N2 production in temperate grasslands: processes, measurements, modelling and mitigating negative impacts. Sci Total Environ 465:173–195. https://doi.org/10.1016/j.scitotenv.2012.11.050
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Clonning: A Laboratory Manual, 2nd Edition. Cold Spring Harbor Laboratory Press, New York, U.S.A., p 1102.
Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-García C, Nissen S (2012) Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. PNAS 109(48):19709–19714. https://doi.org/10.1073/pnas.1211238109
Scala DJ, Kerkhof LJ (1999) Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl Environ Microbiol 65(4):1681–1687. https://doi.org/10.1111/j.1574-6968.1998.tb12979.x
Schmalenberger A, Schwieger F, Tebbe CC (2001) Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl Environ Microbiol 67(8):3557–3563. https://doi.org/10.1128/aem.67.8.3557-3563.2001
Schwieger F, Tebbe CC (1998) A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol 64:4870–4876. https://doi.org/10.1007/springerreference_76320
Schwieger F, Tebbe CC (2000) Effect of Field Inoculation with Sinorhizobium meliloti L33 on the composition of bacterial communities in rhizospheres of a target plant (Medicago sativa) and a non-target plant (Chenopodium album)-linking of 16S rRNA gene-based single-strand conformation polymorphism community profiles to the diversity of cultivated bacteria. Appl Environ Microbiol 66:3556–3565. https://doi.org/10.1128/aem.66.8.3556-3565.2000
Shcherbak I, Millar N, Robertson GP (2014) Global metaanalysis of the nonlinear response of soil nitrous oxide (N2O) emissions to fertilizer nitrogen. PNAS 111(25):9199–9204. https://doi.org/10.1073/pnas.1322434111
Skiba UM, Sheppard LJ, MacDonald J, Fowler D (1998) Some key environmental variables controlling nitrous oxide emissions from agricultural and seminatural soils in Scotland. Atmospheric Environ 32(19):3311-3320. https://doi.org/10.1016/s1352-2310(97)00364-6
Smalla K, Oros-Sichler M, Milling A, Heuer H, Baumgarte S, Becker R, Neuber G, Kropf S, Ulrich A, Tebbe CC (2007) Bacterial diversity of soils assessed by DGGE, T-RFLP and SSCP fingerprints of PCR-amplified 16S rRNA gene fragments: do the different methods provide similar results. J Microbiol Met 69:470–479. https://doi.org/10.1016/j.mimet.2007.02.014
Song B, Leff LG (2005) Identification and characterization of bacterial isolates from the Mir space station. Microbiol Res 160(2):111–117. https://doi.org/10.1016/j.micres.2004.10.005
Stach JE, Bathe S, Clapp JP, Burns RG (2001) PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol Eco 36(2-3):139–151. https://doi.org/10.1016/s0168-6496(01)00130-1
Throbäck IN, Enwall K, Jarvis Å, Hallin S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Eco 49(3):401–417. https://doi.org/10.1016/s0168-6496(04)00147-3
Tsai S, Selvam A, Chang Y, Yang S (2009) Soil bacterial community composition across different topographic sites characterized by 16S rRNA gene clones in the Fushan Forest of Taiwan. Bot Stud 50(1):57–68
Wallenstein MD, Myrold DD, Firestone M, Voytek M (2006) Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecological Appl 16(6):2143-2152. https://doi.org/10.1890/1051-0761(2006)016
White JD, Scott NA (2006) Specific leaf area and nitrogen distribution in New Zealand forests: species independently respond to intercepted light. Forest Ecol Manage 226:319–329. https://doi.org/10.1016/j.foreco.2006.02.001
Wittorf L, Bonilla-Rosso G, Jones CM, Bäckman O, Hulth S, Hallin S (2016) Habitat partitioning of marine benthic denitrifier communities in response to oxygen availability. Environ Microbiol Rep 8(4):486–492. https://doi.org/10.1111/1758-2229.12393
Wolińska A, Górniak D, Zielenkiewicz U, Goryluk-Salmonowicz A, Kuźniar A, Stępniewska Z, Błaszczyk M (2017) Microbial biodiversity in arable soils is affected by agricultural practices. Int Agrophys 31(2):259–271. https://doi.org/10.1515/intag-2016-0040
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Molec Bio Rev 61(4):533–536
Funding
This research has been supported by Ankara University Scientific Research Project Coordination Unit. Project Number: 17L0430004, 2017-2018.
Author information
Authors and Affiliations
Contributions
CA and ESA made a contribution to designing the study. CA was responsible for completing the experiments and data analysis. CA and ESA made a contribution to writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 55 kb)
Rights and permissions
About this article
Cite this article
Avşar, C., Aras, E.S. Community structures and comparison of nosZ and 16S rRNA genes from culturable denitrifying bacteria. Folia Microbiol 65, 497–510 (2020). https://doi.org/10.1007/s12223-019-00754-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-019-00754-8