Abstract
Diffuse large B-cell lymphoma (DLBCL) relapses in approximately 40% of patients following frontline therapy. We reported that STAT6D419 mutations are enriched in relapsed/refractory DLBCL (rrDLBCL) samples, suggesting that JAK/STAT signaling plays a role in therapeutic resistance. We hypothesized that STAT6D419 mutations can improve DLBCL cell survival by reprogramming the microenvironment to sustain STAT6 activation. Thus, we investigated the role of STAT6D419 mutations on DLBCL cell growth and its microenvironment. We found that phospho-STAT6D419N was retained in the nucleus longer than phospho-STAT6WT following IL-4 stimulation, and STAT6D419N recognized a more restricted DNA-consensus sequence than STAT6WT. Upon IL-4 induction, STAT6D419N expression led to a higher magnitude of gene expression changes, but in a more selective list of gene targets compared with STATWT. The most significantly expressed genes induced by STAT6D419N were those implicated in survival, proliferation, migration, and chemotaxis, in particular CCL17. This chemokine, also known as TARC, attracts helper T-cells to the tumor microenvironment, especially in Hodgkin’s lymphoma. To this end, in DLBCL, phospho-STAT6+ rrDLBCL cells had a greater proportion of infiltrating CD4+ T-cells than phospho-STAT6− tumors. Our findings suggest that STAT6D419 mutations in DLBCL lead to cell autonomous changes, enhanced signaling, and altered composition of the tumor microenvironment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common non-Hodgkin’s Lymphoma (NHL), accounting for 30–40% of all worldwide NHL cases [1]. DLBCL is a heterogenous disease, with several gene expression and mutational signatures. Germinal center B-cell (GCB) and activated B-cell (ABC) are the two major molecular subtypes of DLBCL, arising from different steps in B-cell activation and leading to distinct genomic signatures and clinical outcomes [2, 3]. Recently, DLBCL subcategories were re-defined by adding genetic aberrations to gene expression [4,5,6]; however, all DLBCL patients currently receive the same chemo-immunotherapy, R-CHOP: rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone [7]. Although R-CHOP shows significant efficacy, DLBCL relapses in approximately 40% of patients, and rrDLBCL is fatal in 90% of patients [8].
Mechanisms of DLBCL progression following R-CHOP treatment in vivo are unclear, but we hypothesized that mutations enriched in R-CHOP-resistant patients versus newly diagnosed patients play a central role in relapse [9]. Thus, we compared mutations found in R-CHOP-resistant patients with newly diagnosed patients to identify genes with a greater prevalence of mutation at relapse. One such recurrent mutation was in signal transducer and activator of transcription 6 (STAT6) at residue D419, found in 36% of relapsed/refractory GCB-DLBCL (rrDLBCL) and transformed lymphoma (rrTLy) samples [9]. STAT6 is a transcription factor frequently hyperactivated in follicular lymphoma [10], primary mediastinal large B-cell lymphoma (PMBCL) [11] and classical Hodgkin’s lymphoma (cHL) [12]. However, STAT6 hyperactivation in de novo DLBCL is rare, where STAT6 mutation occurs in less than 5% of cases [9]. Despite identification of STAT6 mutations in other lymphomas [10, 13, 14], little is known about how mutation changes its function.
The canonical STAT6 signaling pathway begins with IL-4 or IL-13 binding to their receptors, allowing for receptor autophosphorylation. This phosphorylation creates docking sites for the SH2 domain of STAT6, and recruits JAK proteins that phosphorylate STAT6 at Tyr641. Upon phosphorylation, STAT6 homodimerizes and translocates to the nucleus, where it binds DNA and acts as a transcriptional activator or repressor (reviewed by Goenka and Kaplan [15]). Upon activation, STAT6 has central roles in immune cell functions in normal and pathological conditions [16,17,18,19].
Here, we interrogated steps of the STAT6 signaling cascade in the context of the clinically-relevant D419 mutation to determine how mutated STAT6 function is altered to promote DLBCL cell survival. We asked whether (1) STAT6D419 mutants are constitutively active or have altered kinetics in response to cytokine stimulation, (2) STAT6D419 mutants have altered gene expression program or DNA binding, and (3) any changes in the tumor cell phenotype correlate with D419 mutation. In combination, our data reveals that STAT6D419N has increased nuclear localization upon IL-4 stimulation, where binds to a restricted consensus sequence and promotes increased expression of target genes. Furthermore, STAT6D419 mutant tumor cells secrete more of the chemokine CCL17 upon IL-4 stimulation, and we are the first to provide evidence that this leads to increased CD4+ T-cell infiltration in rrDLBCL patient tumor biopsies.
Materials and methods
Cell culture and transfection
All cell line identities were confirmed by short tandem repeat analyses (Genetica Cell Line Testing, North Carolina). OCI-Ly8, SU-DHL-4, and DB GCB-DLBCL cells were cultured in Roswell Park Memorial Institute (RPMI) media containing 10% fetal bovine serum (FBS) and 0.5% penicillin–streptomycin (P/S). HEK293ft cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS and 0.5% P/S. Transient transfection of HEK293ft cells was performed with Lipofectamine2000, as per manufacturer protocol (see Supplementary reagents and tools table for details of the material used in this work).
Cloning and site-directed mutagenesis
pLX304-eGFP, pLX304-STAT6 (NM_001178079.1), p3xFlag-CMV-14, pCMV-3-IL-4 (NM_000589.3), and LeGO-iT plasmids were purchased. Using site-directed mutagenesis, four STAT6D419 mutated constructs were generated (D419A/G/H/N) in the pLX304 backbone, with a C-terminal V5 tag on each to distinguish between endogenous and overexpressed STAT6 upon transfection or transduction. STAT6D419N was additionally chosen to be cloned into p3xFlag-CMV-14 for immunoprecipitation experiments by amplifying STAT6 with NotI and XbaI restriction sites added, then digesting with NotI and XbaI and ligating into the same restriction sites in the p3xFlag-CMV-14 plasmid.
The pCMV-3-IL-4 plasmid was used to amplify IL-4, with BamHI and NotI restriction sites added, and following digestion with BamHI and NotI restriction enzymes, the IL-4 insert was ligated into LeGO-iT for generation of a stable IL-4 secreting stromal cell line.
Accuracy of cloning was confirmed via sanger sequencing at Genome Quebec.
Generation of STAT6 mutant B-cell lymphoma cell lines
To generate stable STAT6 mutant human GCB-DLBCL cell lines, OCI-Ly8, SU-DHL-4, and DB cells were transduced with pLX304 lentiviruses expressing STAT6WT, STAT6D419A/G/H/N, or empty vector control.
Cellular fractionation and western blotting
Cells were lysed in RIPA buffer to generate whole cell extracts (WCE). To fractionate cells, the cytoplasm was lysed in “Lysis Buffer B” (10 mM Tris at pH 8.4, 140 mM NaCl, 1.5 mM MgCl2, 0.5% IGEPAL, 1 mM DTT), and nuclear protein was further extracted using a detergent stock (3.3% sodium deoxycholate, 6.6% Tween40), RIPA buffer, and harsh sonication.
Protein was separated using 8% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% milk in TBS for 1 h and were incubated with primary antibody overnight at 4℃, followed by secondary antibody for 2 h at room temperature. Membranes were incubated for 1 min with ECL and exposed to film or imaged with a Bio-Rad ChemiDoc. Relative protein expression was calculated via densitometry, using ImageJ.
Chronic IL-4 transwell co-culture model
HEK293ft cells were transduced with LeGO-iT or LeGO-iT-IL-4 lentiviruses, and the resulting IL-4-secreting cells and empty vector control cells were named “293-IL-4” and “293-EV”, respectively. 293-IL-4 cells simulated chronic IL-4 induction using a transwell co-culture model, by seeding 293-EV or 293-IL-4 cells in a 6-well plate at day 1 and seeding OCI-Ly8-pLX304 cells overtop a 0.4 μm pore membrane at day 2. To model acute IL-4 stimulation, OCI-Ly8 cells in co-culture with 293-EV cells were stimulated with 200 pg/mL IL-4 for 3 h.
Immunoprecipitation
Following transient transfection of HEK293ft cells with STAT6WT or D419N-Flag and STAT6WT or D419N-V5, nuclear protein was extracted with Nuclear Complex Co-IP Kit (Active Motif). Protein extracts (250 µg per condition) were pre-cleared using protein G agarose beads. For Flag pull-down, lysates were incubated overnight at 4 °C with M2 beads (Sigma), and for V5 pull-down, lysates were incubated overnight at 4 °C with 2 μg V5 antibody, then were incubated for 2 h at room temperature with magnetic dynabeads. For both pulldowns, beads were washed 3 times with lysis buffer following immunoprecipitation, then were boiled in 2 × Laemmli buffer prior to western blotting.
RNA sequencing and ingenuity pathway analysis
RNA was extracted using the Absolutely Total RNA Purification Kit (Agilent). RNA sequencing was performed at Génome Québec (see supplemental methods for RNA sequencing analysis and Ingenuity Pathway Analysis).
qPCR
RNA extracted using the E.Z.N.A. Total RNA Kit I (Omega Bio-Tek) was used for RT-PCR. cDNA was diluted 1/10 for qPCR with GoTaq qPCR master mix (Promega), using 100 nM primers. Transcript levels were normalized to three reference genes (TBP, RPLP0, and HPRT1), and fold change was calculated relative to empty vector control using the ΔΔCt method.
Molecular modelling
Using the STAT6 DNA complex structure (PDBID: 4Y5W), loop modelling techniques were applied to predict the starting conformation of undefined loops (247–252, 324–336, 395–399) and were energy minimized. The system was prepared for molecular dynamics (MD) simulations by protonating all residues and correcting charge and hybridization states when needed. D419 residues were manually modified in both STAT6 monomer sequences using UCSF Chimera and new side chains were energy minimized.
Molecular dynamics simulations were conducted using AMBER20 software. Tleap module was used for neutralizing and solvating the system, defining a 10 Å truncated octahedron TIP3P water box. Solvent was energy minimized and heated to 300 K (200 ps, NTP) keeping the rest of the system restrained (defining 10 kcal/mol force constant). The whole system was therefore energy minimized to reduce the intermolecular clashes. Once the energy was minimized, non-water molecules were fixed again by applying positional restraints (10 kcal/mol) and the system was heated to 300 K (100 ps, NVT). A 100 ps NPT density equilibration stage, keeping the restraints, was performed before a progressive reduction of the restraints with four NVT equilibration stages (10, 8, 5, 1 kcal/mol). A final 100 ps NPT density equilibration without restraints was conducted before a 30 ns NPT production stage.
For umbrella sampling simulations, 50 independent molecular simulations (5 ns/window) were performed using AMBER20 software defining a movement of 1 Å in each window (force constant, k = 4 kcal/mol/Å). The Potential of Mean Force was calculated using WHAM (defining 1·10–5 tolerance).
Binding motif analysis
STAT6 DNA-binding sequences were identified with the Gene Transcription Regulation Database (GTRD) [20]. The Analysis of Motif Enrichment (AME) [21] tool of the MEME Suite 5.4.1 was used to identify the presence of the STAT6 canonical consensus motif and the Multiple Em for Motif Enrichment (MEME) [22] tool was used to identify novel consensus motifs (see supplementary materials and methods).
Enzyme-linked immunosorbent assay (ELISA)
293-EV or 293-IL-4 cells were seeded in a 6-well plate at day 1, and OCI-Ly8, DB, or SU-DHL-4 cells were seeded overtop a 0.4 μm pore membrane at day 2. On day 5, media was collected and snap frozen. CCL17 was quantified in the conditioned media using R&D human CCL17 DuoSet ELISA. Optical density (OD) values were calculated by subtracting the OD 540 nm from the OD 450 nm.
CCL17-reporter assay
293-EV or 293-IL-4 cells were transfected with the CCL17-reporter plasmid containing the STAT6 responsive element upstream of the Firefly luciferase (fLuc) gene [23] (a kind gift from Dr. Daniel Hebenstreit, University of Warwick), a Renilla luciferase (rLuc) control plasmid (Promega), and either pLX304-EV, pLX304-STAT6WT, or pLX304-STAT6D419N. Twenty-four hours later, fLuc and rLuc activities were measured with a Dual-Luciferase assay kit (Promega). Relative luminescence units (RLU) were calculated by normalizing fLuc to rLuc.
Patient data
The cohort used for microarray analysis and immunohistochemistry was previously described [9], and consisted of samples obtained prospectively from patients treated in Montreal QC and enrolled in the QCROC-2 clinical trial NCT01238692 or obtained from the Leukaemia Cell Bank of Quebec-Lymphoma axis. Patients provided informed consent for the clinical trial or for storage of samples in the databank. This project was approved by the research ethics board at the Jewish General Hospital, in accordance with the declaration of Helsinki (biobanking approval number 11-046 and STAT6 project 15-036).
An additional validation cohort was obtained from BC Cancer and consisted of matched transcriptomic and genomic data from de novo DLBCL patients [24,25,26] (see supplementary materials and methods for details).
Immunohistochemistry
Immunohistochemistry (IHC) was performed using a Discovery XT Autostainer, with primary antibody incubation for 30 min at 37 °C. Tissues were counterstained with Hematoxylin, blued with Bluing Reagent, dehydrated through graded alcohols, cleared in xylene, and mounted. Sections were scanned at 20× using an Aperio AT Turbo Scanner. Staining intensity was scored by a blinded pathologist.
Statistical analysis
Data was analyzed by unpaired Student’s t test, 1way ANOVA, or 2way ANOVA with Tukey’s post-hoc analysis. Sample size and statistical test are indicated in figure legends, where applicable.
Data availability statement
Raw and processed RNA sequencing data are available at GEO accession number GSE220762.
Results
STAT6 D419 mutation neither increases the proliferative rate of GCB-type DLBCL cells nor STAT6 phosphorylation in response to IL-4
The enrichment of STAT6D419 mutations at relapse suggests they contribute to cell survival in DLBCL. Indeed, knock-down of STAT6 in DLBCL, PMBCL, cHL, and FL reduces cell viability in the absence of IL-4, which suggests that STAT6 has tonic survival functions [13, 27, 28]. To test this, we generated OCI-Ly8 [29], SU-DHL-4 [30], and DB [31] cell lines mimicking the heterozygous expression of mutations found in patient biopsies by overexpressing STAT6WT or STAT6D419 mutants (Fig. 1A). Each of these cell lines are known to be derived from GCB-type DLBCL, and each of their mutational profiles further suggests that they are of the EZB subtype, as defined by Wright et al. [4] and characterized by mutations in genes such as EZH2, CREBBP, and KMT2D. STAT6 mutations are typically associated with the EZB subtype, and our patient data further asserts that STAT6 mutant rrDLBCL is of the EZB subtype (Fig. S1). Overexpression of STAT6WT or STAT6D419A/G/H/N had no effect on DLBCL growth, as compared to empty vector controls (Figs. 1B, S2A). In addition, expression of STAT6D419 mutation alone had no impact on cellular response to any of the individual components of R-CHOP therapy (Fig. S2C). Together, these results indicate that STAT6D419 mutant cells do not have enhanced proliferation or increased therapeutic resistance in the absence of cytokine stimulation.
STAT6D419 cells have similar response to IL-4 as STAT6WT cells. A Schematic demonstrating the generation of STAT6 mutant DLBCL cell lines. B Growth curve of OCI-Ly8-pLX304 cell lines, expressing empty vector, STAT6WT, or STAT6D419A/G/H/N. Cells were counted once every 24 h for 96 h using a haemocytometer and trypan blue exclusion. Data shown are 4 biological replicates, consisting of 3 technical replicates each. C Representative western blot of 3 biological replicates demonstrating OCI-Ly8 sensitivity to increasing concentrations of IL-4 for 30 min. (S.E: Short exposure; L.E: Long exposure). D Representative western blot of 3 biological replicates demonstrating OCI-Ly8 time course of 200 pg/mL IL-4 stimulation. E Representative western blot of 3 biological replicates demonstrating OCI-Ly8 200 pg/mL IL-4 wash off time course. F Schematic illustrating the OCI-Ly8 and 293-EV/IL-4 co-culture system. G Western blot demonstrating that co-culture with 293-IL-4 induces sustained STAT6 phosphorylation. H Growth curve of OCI-Ly8 cell lines co-cultured with 293-EV or 293-IL-4 for 72 h. Data shown are 4 biological replicates, consisting of 2 technical replicates each. In all cases, bar graphs show densitometry of phospho-STAT6 expression, normalized to GAPDH or β-actin loading control expression
We observed enriched phospho-STAT6 staining in DLBCL patient biopsies with a STAT6D419 mutation [9], suggesting that these mutations lead to a constitutively active STAT6, altered kinetics of response to cytokine, or increased cytokine signaling from the microenvironment. To test the first two of these possibilities, phospho-STAT6 was assessed with increasing concentrations of IL-4. STAT6 was not phosphorylated in the absence of IL-4 (Fig. 1C), and each cell line had similar STAT6 phosphorylation in response to different concentrations of IL-4 (Fig. 1C). We next assessed whether STAT6D419N mutation changed the kinetics of STAT6 phosphorylation. IL-4-induced phospho-STAT6 expression peaked between 30 min and 1 h, and gradually declined to background levels by 24 h in all cell lines (Figs. 1D, S2B). Furthermore, when cells were treated with IL-4 for 3 h and media was removed, the loss of phospho-STAT6 was similar between cell lines (Fig. 1E). These findings indicate that STAT6D419N is not constitutively active nor is STAT6D419N phosphorylation altered in response to IL-4. Of note, we found that exogenous expression of STAT6WT and STAT6D419N had no impact on total STAT6 expression levels in OCI-Ly8 and SU-DHL-4 cells (Figs. 1C, S2), suggesting that there is a buffering mechanism that keeps total STAT6 protein levels static.
Our data demonstrate that cytokine stimulation is required for mutant STAT6 phosphorylation. To test whether D419 mutation alters cell growth in the presence of sustained cytokine signaling, we developed a co-culture model wherein DLBCL cells are cultured in a transwell above a stromal cell line that constitutively produces IL-4 (293-IL-4; Fig. 1F), leading to sustained STAT6 phosphorylation (Fig. 1G). Using this co-culture system, OCI-Ly8, DB, and SU-DHL-4 expressing STAT6WT or STAT6D419A/G/H/N cells have a similar proliferative rate with and without IL-4 induction (Figs. 1H, S2D-F). Thus, IL-4 does not enhance the proliferative rate of STAT6D419 cells in vitro.
STATD419N can heterodimerize with STAT6WT.
In the STAT6 canonical signaling cascade, STAT6 dimerizes upon phosphorylation, and translocates to the nucleus [15]. This dimerization step is critical within the STAT6 activation cascade; thus, we questioned whether D419N mutation impacts the ability of STAT6 to form dimers and whether STAT6D419N could dimerize with STAT6WT. To test the ability of STAT6WT and STAT6D419N to homo- and heterodimerize, we used co-immunoprecipitation (co-IP) in HEK293ft cells transiently expressing tagged STAT6WT or STAT6D419N capable of increasing phospho-STAT6 in response to IL-4 (Fig. S3). We transfected Flag-tagged and V5-tagged STAT6WT or STAT6D419N and stimulated cells for 1 h with IL-4. By immunoprecipitating with anti-Flag and probing for V5, and the reciprocal, we found that STAT6WT and STAT6D419N can both homodimerize and heterodimerize (Fig. 2A). Interestingly, STAT6WT-STAT6D419N heterodimers and STAT6D419N homodimers were immunoprecipitated from nuclear extracts more readily upon IL-4 stimulation, as compared to STAT6WT homodimers (Fig. 2B). The latter suggests an increased nuclear localization of STAT6 dimers when one or both monomers contain the D419N mutation.
Increased presence of STAT6D419N hetero- and homodimers in the nucleus upon IL-4 stimulation. A Representative western blot of 3 biological replicates from co-IP experiments. Lysates used for co-IP were from nuclear fractions. Labelling indicates transfection conditions. “Input” immunoblots are from whole cell extracts, to confirm equal transfection conditions. B Densitometry of Flag IP. Band intensity was normalized to input control, and then represented as fold change in intensity from the STAT6WT homodimerization -IL-4 condition. C Representative western blot of 3 biological replicates from OCI-Ly8-EV, OCI-Ly8-STAT6WT, and OCI-Ly8-STAT6D419N cellular fractions stimulated with 200 pg/mL IL-4 over 6 h. The nuclear fraction shows phospho-STAT6 nuclear accumulation with D419N mutation. D Densitometry of nuclear phospho-STAT6 expression, normalized to H1 loading control expression and expressed as fold change from the EV without IL-4. E, F Representative western blots of 3 biological replicates from cytoplasmic and nuclear cell fractions of transfected 293-EV cells stimulated with IL-4 for 3 h (acute stimulation; E) and transfected 293-IL-4 cells (chronic stimulation; F). Labelling indicates transfection conditions. Bar graphs show densitometry of p-STAT6 expression, normalized to GAPDH or H3 loading control. Bars in the graph are presented corresponding to western blot loading. Statistics on all bar graphs are 2-way ANOVA with multiple comparison (*p < 0.05, ** = p < 0.01, *** = p < 0.005, **** = p < 0.001)
STAT6D419N displays increased nuclear translocation.
Next, we assessed STAT6D419N phosphorylation kinetics in cellular fractions following IL-4 stimulation in OCI-Ly8 cell lines. Cells were stimulated with IL-4 for 1, 3, and 6 h, and phospho-STAT6 expression was assessed in whole cell, cytoplasmic, and nuclear protein extracts. In whole cell extracts, phospho-STAT6 induction by IL-4 was similar between STAT6WT and STAT6D419N, consistent with our initial findings (Figs. 1D, 2C). Likewise, within the cytoplasmic fraction, phospho-STAT6 was similar between STAT6WT and STAT6D419 mutant cells. However, within the nuclear fraction, STAT6D419N displayed increased phosphorylation in response to IL-4 (Fig. 2C–D). Similar results were obtained with SU-DHL-4 cells (Fig. S4A) and when STAT6D419N mutant OCI-Ly8 cells were stimulated with IL-13 (Fig. S4B). We also confirmed HEK293ft cells transiently transfected with STAT6D419N have increased nuclear expression both upon acute (3 h; Fig. 2E) and chronic (72 h; Fig. 2F) exposure to IL-4. Thus, STAT6D419 mutants have increased nuclear expression following cytokine activation, concordant with patient data showing strong nuclear phospho-STAT6 staining [9].
STAT6D419N upregulates a more stringent set of genes with increased magnitude
We next hypothesized that the increased nuclear expression of STAT6D419N might lead to altered transcription factor activity and thereby, altered gene expression. To assess differential gene expression signatures, we extracted RNA from OCI-Ly8 cells expressing either STAT6WT or STAT6D419N grown in co-culture with 293-EV or 293-IL-4 cells for 3 h (acute stimulation) or 72 h (chronic stimulation) and performed RNA sequencing. In the absence of IL-4, gene expression between STAT6WT and STAT6D419N was similar and clustered by exposure time rather than genotype by hierarchical clustering (Fig. 3A). Only 4 genes were differentially expressed in the absence of IL-4 between STAT6WT and STAT6D419N cells, including PRKCB, IL1RAPL1, ZNF492, and RN7SL396P. We compared the IL-4-dependent differentially expressed genes (DEGs) between STAT6WT and STAT6D419N cells and identified only 84 DEGs after acute IL-4 treatment and 2,185 DEGs after 72 h (Fig. 3B). Of note, the DEGs which distinguished STAT6WT and STAT6D419N cells following 72 h IL-4 treatment were often upregulated in STAT6D419N as compared to STAT6WT (Fig. 3C). However, we also found that IL-4 induced DEGs are often shared by both cell lines (35% in acute and 42% in chronic exposure; Fig. 3B), and include the STAT6 transcriptional targets FCER2, IL4R, AMICA1, SOCS1, and DPP4. Using qPCR, we validated the expression of these genes, as well as some novel genes found to be upregulated by IL-4 induction, such as NR4A3, LTBP1, and MOB3C. Each of these genes were upregulated in STAT6WT and STAT6D419N cells upon chronic IL-4 stimulation, and gene expression of SOCS1, DPP4, and NR4A3 was induced more in STAT6D419N cells than in STAT6WT cells (Fig. 3D). Furthermore, expression of each of these genes depends on sustained IL-4 signaling, as transcript levels decline when IL-4 is removed from cell culture media, leading to pre-stimulation levels by 6–12 h post-IL-4 wash off (Fig. S5).
STAT6D419N upregulates a restricted set of genes with increased magnitude. A Dendrogram demonstrating that OCI-Ly8-STAT6WT and OCI-Ly8-STAT6D419N groups cluster by IL-4 treatment time. B Venn diagram showing the number of DEGs upon 3 h and 72 h IL-4 treatment. Upon IL-4 treatment, STAT6WT has more unique DEGs than STAT6D419N. C Heatmap showing gene expression differences between STAT6WT and STAT6D419N cells, upon 72 h IL-4 treatment. Blue or red intensity represents Z-score. D qPCR validations of RNAseq data demonstrate that STAT6D419N upregulates transcription of gene targets with increased magnitude. Data consist of 4 biological replicates, with 3 technical replicates each (2way ANOVA; *p < 0.05, ** = p < 0.01, *** = p < 0.005, **** = p < 0.001)
Interestingly, there were more DEGs exclusive to STAT6WT than STAT6D419N cells at both time points. After chronic IL-4 exposure, there were 5 × more DEGs selective for STATWT (1062 in WT vs 205 in D419N; Fig. 3B). These results were surprising, given that STAT6D419N demonstrated enhanced nuclear localization. Together, our RNA sequencing and qPCR data demonstrate that STAT6D419N upregulates the expression of a more selective gene set, but with increased magnitude, upon IL-4 stimulation.
STAT6D419 mutation does not uniformly impact DNA-binding dynamics.
The D419 residue locates along the major groove of the protein-DNA-binding interface, and it has therefore been previously predicted that mutation of the D419 residue may influence DNA binding by STAT6 [10, 13]. Thus, we next questioned how D419 mutation of STAT6 might alter DNA interactions. When MD simulations were used to model the molecular structure of endogenous DNA-bound STAT6WT (Figs. 4A, S6A) and STAT6D419N (Fig. 4B), no remarkable differences in the backbone geometry were observed. Likewise, MD were also used to describe STAT6 DNA binding by monitoring the evolution of three distances: first, the distance between both SH2 domains; second, the distance between D419 residue and DNA centres of mass (chain A); and third, the distance with DNA chain B. No major differences were found for STAT6WT and STAT6D419N systems (Figs. 4C, S6B, C). Once bound, STAT6WT and STAT6D419N showed similar ability to unravel the DNA structure, as illustrated by the predicted number of hydrogen bonds between base pairs and the base paired ratio (Figs. 4D, S6D). Furthermore, umbrella sampling simulations were used to model STAT6 release from the DNA. Starting from the stable DNA-bound conformation, the DNA structure was pulled out of the STAT6 structure, by increasing the distance from SH2-SH2 and DNA from 30 to 80 Å, until the DNA was released from the protein. These simulations found that STAT6 released from DNA around 50 Å, and D419N mutation did not induce a change in the free energy of binding (Figs. 4E, S6E). While STAT6D419N showed no remarkable differences in DNA-binding dynamics from STAT6WT, STAT6D419A and STAT6D419G did have significantly changed DNA binding affinity (Fig. S6), with STAT6D419A and STAT6D419G showing greater ability to unravel the DNA secondary structure, and STAT6D419G additionally showing an increase in the free energy required for DNA unbinding. These findings indicate that D419 mutation does not uniformly change STAT6 DNA interactions, and that increased DNA-binding affinity does not necessarily underlie the increased nuclear presence of STAT6 mutants or the increased transcription of STAT6 target genes.
STAT6D419N recognizes an altered DNA-consensus motif. A, B Ribbon diagrams showing the DNA-bound conformation of STAT6WT and STAT6D419N. C Molecular dynamic simulations showing STAT6 DNA-binding dynamics. SH2-SH2 shows the distance between SH2 domains measured between their centre of mass, A-DNA shows the distance between one of the D419 residues and the DNA centre of mass (chain A, resID 291), and B-DNA shows the distance between the other D419 residue and the DNA centre of mass (chain B, resID 814), during the process of STAT6WT or STAT6D419N binding to DNA. D Evolution of the number of DNA intermolecular hydrogen bonds and the paired bases ratio along the MD simulation. When purple and orange lines jump from one limit to the other, it denotes a higher ratio of frames without H-bonds and a lower BP-ratio. E Graphical representation of the umbrella sampling study, and Potential of Mean Force (PMF) obtained after umbrella sampling simulations. The distance between SH2-SH2 and DNA was considered as the collective variable under study. Fifty independent simulations were conducted for each STAT6 DNA complex to explore all the molecular states when changing the collective variable from 30 to 80 Å. The overlap in density plots along the coordinate assure the thermodynamic reliability of the study. PMF approximates the free energy landscape when following a coordinate of interest (a.k.a. collective variable). On the right, the PMF involving the DNA unbinding process is shown. F Venn diagram showing the number of identified STAT6-binding sequences identified within proximity of genes upregulated by STAT6WT or STAT6D419N upon 3 h IL-4 treatment. G STAT6WT and STAT6D419N novel consensus motifs, as identified with MEME using STAT6 DNA-binding sequences obtained from GTRD
STAT6D419N recognizes a restricted DNA-consensus sequence.
STAT6D419N regulates the expression of a more stringent set of genes than STAT6WT, so we next questioned whether there was a difference in DNA-binding motifs recognized by STAT6WT and STAT6D419N. Genes upregulated upon 3 h of IL-4 stimulation are likely to be primary targets of STAT6 transcription factor activity, thus, this dataset was used for this analysis. ChIP-seq data compiled by GTRD [20] were used to identify STAT6 DNA-binding sequences upstream and downstream of genes upregulated in STAT6WT only (72 sequences), STAT6D419N only (23 sequences), and in both STAT6WT and STAT6D419N (58 sequences); thus 130 STAT6-binding sequences were associated with STAT6WT, and 81 STAT6-binding sequences were associated with STAT6D419N (Fig. 4F). Within these STAT6-binding sequences, the canonical palindromic STAT6 5′-TTC(N)3…4GAA-3′ consensus motif [32] was identified in 10% of STAT6WT-associated sequences (13/130; Evalue = 9.29e−1), and in 8.6% of STAT6WT-associated sequences (7/81; Evalue = 6.14e−1), serving as an internal validation of this methodology, but also suggesting the canonical STAT6-binding consensus motif is not significantly enriched within our dataset. However, a novel DNA-consensus sequence motif was identified within our dataset (Fig. 4G) at 70 sites with an E value of 1.5e−014 in STAT6WT-associated binding sequences and at 59 sites with an E value of 5.9e−005 in STAT6D419N-associated binding sequences. Interestingly, this binding motif is restricted to a guanine residue at positions 3 and 6 in sequences regulated by STAT6D419N (G-NN-G motif), while the residues at positions 3 and 6 are more variable in sequences regulated by STAT6WT. These results suggest that the binding of STAT6D419N to DNA is restricted to a more specific sequence than STAT6WT.
STAT6D419 mutants upregulates CCL17 expression to recruit CD4 + T-Cells
To further understand the DEGs selective to STAT6WT or STAT6D419N, we performed pathway analyses of the genes significantly altered in STAT6WT or STAT6D419N OCI-Ly8 cells upon chronic IL-4 induction. We found that STAT6D419N cells have increased activation of pathways that have genes related to cell viability and survival, and less activation of pathways that have genes related to organismal death and cell morbidity/mortality as compared to STAT6WT, suggesting that STAT6D419N cells have improved survival in response to IL-4. Notably, we found that expression of STAT6D419A/G/H/N does not confer a proliferative advantage in GCB-DBCL cell lines, although these cell lines are culture-adapted and highly proliferative, potentially masking a mutation-induced phenotype. STAT6D419N cells also have increased activation in pathways related to chemotaxis and cell migration, including cell movement and migration of mononuclear cells and lymphocytes (Fig. 5A; Table 1). Together, these data indicate that STAT6D419N cells have a survival advantage in response to IL-4 stimulation and may have increased capability to attract or migrate to specific immune cells that could be IL-4 secreting.
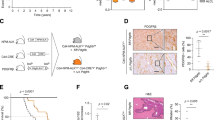
STAT6D419 mutation results in increased CCL17 secretion and increased invasion of CD4 T-Cells. A IPA analysis of RNAseq data showing STAT6D419N cells have enrichment in pathways related to Cell Viability and Cell Migration and Chemotaxis. B qPCR showing CCL17 transcription is increased in STAT6D419N cells upon IL-4 stimulation. Data consist of 4 biological replicates in technical triplicate (2-way ANOVA; ****p < 0.001). C ELISA showing CCL17 secretion is increased in STAT6D419N cells compared to STAT6WT cells upon IL-4 stimulation. Data consists of 3 biological replicates in technical triplicate (2-way ANOVA; **p < 0.01). D CCL17-reporter luciferase assay showing CCL17 transcription is increased 30-fold when STAT6D419N is expressed in 293-IL-4 cells. (RLU: relative luminescence units). E CCL17 expression obtained from gene microarray in GCB rrDLBCL patients who had negative or positive phospho-STAT6 staining (two-tailed unpaired t test; *p < 0.05). Stars indicate the presence of a STAT6D419 mutation. F CCL17 expression obtained from a compendium of RNA sequencing of 598 de novo DLBCL patients and 1 transformed lymphoma patient, comparing CCL17 expression in GCB STAT6WT patients with CCL17 expression in STAT6 non-D419 mutant patients (STAT6Mut) and STAT6D419 mutant patients (1-way ANOVA; **p < 0.01). Of the STAT6Mut patients, 4 were GCB and 2 were ABC. Of the STAT6D419 mutant patients, 9 were GCB and 1 was unclassified G–I CD3, CD4, and CD8 staining in GCB rrDLBCL patient biopsies. CD3, CD4, or CD8 staining intensity score was compared between phospho-STAT6 negative and phospho-STAT6 positive patients (scored by a blinded pathologist; 1 = negative staining, 2 = weak staining, 3 = moderate staining, 4 = strong staining). Scale bars below each image indicate 100 µm. Stars indicate the presence of a STAT6D419 mutation. J CCL17 expression obtained from gene microarray in GCB rrDLBCL patients who had low CD4 staining (score 1–2), or high CD4 staining (score 3–4; unpaired t test; ***p < 0.005). Stars indicate the presence of a STAT6D419 mutation
STAT6D419 mutant rrDLBCL patient biopsies have increased phospho-STAT6 staining and an increased STAT6 expression signature [9], suggesting that STAT6D419-mutants are able to recruit IL-4 secreting cells to maintain an activated state. This aligns with the pathway analyses showing enrichment in chemotactic signals. One interesting candidate from our dataset and a known STAT6 transcriptional target is CCL17 (aka TARC), a chemokine that can attract various CD4+ T-cells, such as Th2 cells and Tregs. CCL17 is upregulated in human FL samples that express mutated STAT6 [10, 13], but this has not been shown in STAT6 mutant DLBCL cell lines. To this end, qPCR determined that IL-4-induced CCL17 mRNA expression is significantly upregulated by STAT6D419A/G/H/N as compared to control, in OCI-Ly8, SU-DHL-4, and DB cells (Figs. 5B, S7A-C). In addition, CCL17 was more highly secreted by STAT6D419A/G/H/N than STAT6WT cells following chronic IL-4 induction (Figs. 5C, S7D-F). Using a CCL17-reporter luciferase assay in 293-EV and 293-IL-4 cells, we confirmed the upregulation of CCL17 by STAT6D419N was indeed transcriptional (Fig. 5D).
We assessed whether this observation could be translated to clinical samples, using our previously published gene microarray data [9] from CD19 + cells selected from rrDLBCL biopsies that were also annotated for phospho-STAT6 expression, indicating STAT6 activation by IL-4. Patients were stratified into “phospho-STAT6 positive” and “phospho-STAT6 negative” groups, with the “phospho-STAT6 positive” group enriched in STAT6D419 mutations, but also including mutations in other members of the STAT6 signaling axis (i.e., IL4R, SOCS1, and JAK2). This allowed us to expand our analyses to all rrDLBCL samples with the same phenotype. We found that phospho-STAT6 positive rrDLBCL tumors had increased CCL17 mRNA expression (Fig. 5E). In addition, using an independent cohort from BC Cancer, where phospho-STAT6 histology data was not available, CCL17 expression was compared in STAT6WT and STAT6D419 mutant patient samples. Although these samples were obtained from de novo and not relapsed/refractory patients, CCL17 was still found to be significantly increased when a D419 mutation was present (Fig. 5F). Furthermore, in this dataset, CCL17 expression was not found to be increased in patients with a non-D419 STAT6 mutation, indicating a unique role for the mutated D419 residue.
Given the role of secreted CCL17 as a chemoattractant, we hypothesized that the surrounding microenvironment of phospho-STAT6 positive tumors might be enriched in T-cells. Using serial sections from rrDLBCL biopsies, we assessed the presence of CD3, CD4, and CD8 antigens with IHC, and found that phospho-STAT6 positive rrDLBCL tumors have a significantly higher proportion of CD3+ T-cells (Fig. 5G). Half of phospho-STAT6 positive tumors had a high percentage (25–50%) of CD4+ T-cells, and of the 3 phospho-STAT6+/CD4High samples, 2 had a STAT6D419 mutation (Fig. 5H). Moreover, even the phospho-STAT6 negative samples with a STAT6D419 mutation were found to have high infiltration of CD4+ T-cells. Interestingly, there were no significant differences in the presence of CD8+ T-cells when samples were stratified by phospho-STAT6 status (Fig. 5I). However, when we stratified the data by STAT6 mutation, CD8+ T-cells were significantly increased in STAT6 mutant biopsies, in addition to the CD4+ T-cells (Fig. S8). Finally, to establish a link between CCL17 in rrDLBCL tumor cells and CD4+ T-cell infiltration, CCL17 mRNA expression was compared between CD4High and CD4Low samples, and there was a striking increase in tumor cell CCL17 mRNA expression in samples with CD4High staining (Fig. 5J). Together, these results indicate that STAT6 mutant rrDLBCL cells remodel their tumor microenvironment (TME) via the secretion of the chemokine CCL17 to attract CD4+ T-cells, although the subtype of CD4+ T-cells which were recruited were not determined in this study.
Discussion
STAT6 mutations are enriched at relapse in DLBCL and localize to the D419 hotspot within the DNA-binding domain [9], implying that mutations in the STAT6D419 DNA-binding domain are functionally significant in rrDLBCL. Given that relapse in DLBCL is considerable, and the mechanisms behind tumor growth post-R-CHOP are poorly elucidated, understanding how STAT6D419 mutations contribute to DLBCL relapse is an important clinical matter. Thus, our study used exogenously expressed STAT6WT and STAT6D419A/G/H/N in 3 different GCB-DLBCL cell lines to examine how STAT6 signaling is altered by D419 mutation. Upon IL-4 stimulation, STAT6D419N has increased nuclear localization, perhaps recognizing a restricted DNA-consensus sequence, and resulting in a more stringent gene expression profile. Although molecular modeling did not indicate altered DNA binding, we only modeled the canonical STAT6 consensus sequence and did not account for altered nuclear protein expression. In addition to DNA binding, we hypothesize that increased nuclear localization may be due to 1) altered nuclear import or export [33]; or 2) altered protein–protein interactions contributing to nuclear retention.
Yildiz et al., previously observed that STAT6D419G mutant cells exhibited nuclear STAT6 expression independent of IL-4 stimulation, but phosphorylation of STAT6 is still cytokine dependent [13]. Also, Yildiz et al. did not find global basal transcriptional differences compared to STAT6WT, which is consistent with our findings, suggesting that nuclear presence alone does not stimulate transactivation activity. Furthermore, Mentz et al. reported that STAT6D419H mutations can be characterized as gain-of-function, but the mutant STAT6 protein still requires IL-4 stimulation for transactivation [10]. We have extended these data, showing that while mutant STAT6 hyperactivates some targets, STAT6D419N has a more restricted set of target genes than the wild-type protein. STAT6 hyperactivation has been previously attributed to autocrine IL-13 production [34], JAK2 amplification [11], and a combination of autocrine IL-4/IL-13 production and overall enrichment of microenvironmental cytokines [12, 35,36,37,38]. In our hands, STAT6D419N does not have increased nuclear residency in the absence of IL-4, nor do mutant GCB-DLBCL cells display evidence of autocrine IL-4/IL-13 induction or JAK2 co-mutation or amplification [9]. Thus, we predict that STAT6 activation in STAT6 mutant rrDLBCL is due to cytokine secretion from a constituent of the TME.
STAT6D419 mutations have been previously investigated in the context of FL, which is a slow-growing lymphoma that relies on the TME for cancer cell survival [39]. While it has been shown that STAT6D419 mutation leads to upregulated CCL17 and CCL22 in FL patient samples [10], our group is the first to characterize STAT6D419 mutations in the context of rrDLBCL. rrDLBCL is the most deadly DLBCL, and is often characterized by an immune “depleted” TME [40]. However, STAT6 mutations in DLBCL are associated with a GC-type TME that is enriched in various CD4 T-cells [40]. To this end, in our rrDLBCL patient samples, phospho-STAT6 positivity and STAT6D419 mutation correlates with increased CCL17 expression. CCL17 is a known STAT6 transcriptional target [41] and chemoattractant that recruits various CCR4 + immune cells—such as CD8+ or CD4+ T-cells, and macrophages—in murine and canine cancer models, in addition to human tumors [42,43,44,45]. While CCL17 is not normally secreted by B cells, it has previously been reported that expression can be induced [46, 47]. Indeed, in our study, IL-4 stimulation of STAT6 mutant cells induced an upregulation of CCL17 expression, and furthermore, in patient samples, increased CCL17 expression correlated with increased invasion of CD4+ T-cells. Therefore, we postulate that STAT6D419-induced-expression of CCL17 may recruit CD4+ T-cells capable of producing IL-4/IL-13, including Th2, Tfh, and Treg subsets, to maintain STAT6 activation (Fig. 6: Schematic Summary). Furthermore, we predict that there may be additional cascading impacts on the TME. For instance, if the CD4 + T-cell infiltrate is indeed IL-4/IL-13 secreting, this may have functional consequences on the non-tumor cells of the TME.
Schematic Summary of Data. 1. In STAT6WT and STAT6D419N cells, IL-4 is required for STAT6 activation. 2. Upon activation with IL-4, both STAT6WT and STAT6D419N will dimerize and translocate to the nucleus. 3. However, STAT6D419N has increased nuclear retention as compared to STAT6WT and recognizes an altered DNA-consensus sequence. 4. STAT6D419N has increased transcription of select gene targets, such as CCL17. 5. Increased transcription of CCL17 by STAT6D419N leads to increased production and secretion of CCL17, which functions to recruit CD4+ T-cells
We speculated that the STAT6 mutant protein plays a role in therapeutic resistance, however, we found that STAT6D419A/G/H/N mutations in GCB-DLBCL cells have no impact on cellular response to the individual components of R-CHOP. Thus, STAT6 mutations don’t directly cause resistance, but there is ample evidence that the TME is prognostic in DLBCL [40, 48, 49]. To this end, further investigation is warranted to determine if therapeutic resistance is associated with STAT6D419-induced microenvironmental remodeling. From a therapeutic angle, knowing how the tumor influences the surrounding immune cells is important, as cellular therapies, such as CAR-T [50, 51], are being increasingly used to treat rrDLBCL. Future studies may examine if STAT6 mutations affect responses to these novel therapies, by using immunocompetent mouse models that allow for the careful dissection and manipulation of the TME. Overall, our data show that tumor cell autonomous changes in STAT6 result in microenvironmental changes, and we believe further investigation could have an impact on informing standard of care for rrDLBCL patients, and potentially other B-cell malignancies identified with STAT6 mutations [10, 13, 14], by identifying novel therapeutic susceptibilities.
Data availability
All data is available upon request.
References
Susanibar-Adaniya S, Barta SK. 2021 Update on diffuse large B cell lymphoma: a review of current data and potential applications on risk stratification and management. Am J Hematol. 2021;96(5):617–29.
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–11.
Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–47.
Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37(4):551-68.e14.
Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396–407.
Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679–90.
Coiffier B, Lepage E, Brière J, Herbrecht R, Tilly H, Bouabdallah R, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(4):235–42.
Sehn LH, Gascoyne RD. Diffuse large B-cell lymphoma: optimizing outcome in the context of clinical and biologic heterogeneity. Blood. 2015;125(1):22–32.
Morin RD, Assouline S, Alcaide M, Mohajeri A, Johnston RL, Chong L, et al. Genetic landscapes of relapsed and refractory diffuse large B-cell lymphomas. Clin Cancer Res. 2016;22(9):2290–300.
Mentz M, Keay W, Strobl CD, Antoniolli M, Adolph L, Heide M, et al. PARP14 is a novel target in STAT6 mutant follicular lymphoma. Leukemia. 2022;36(9):2281–92.
Guiter C, Dusanter-Fourt I, Copie-Bergman C, Boulland ML, Le Gouvello S, Gaulard P, et al. Constitutive STAT6 activation in primary mediastinal large B-cell lymphoma. Blood. 2004;104(2):543–9.
Van Slambrouck C, Huh J, Suh C, Song JY, Menon MP, Sohani AR, et al. Diagnostic utility of STAT6(YE361) expression in classical Hodgkin lymphoma and related entities. Mod Pathol. 2020;33(5):834–45.
Yildiz M, Li H, Bernard D, Amin NA, Ouillette P, Jones S, et al. Activating STAT6 mutations in follicular lymphoma. Blood. 2015;125(4):668–79.
Ritz O, Guiter C, Castellano F, Dorsch K, Melzner J, Jais JP, et al. Recurrent mutations of the STAT6 DNA binding domain in primary mediastinal B-cell lymphoma. Blood. 2009;114(6):1236–42.
Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunol Res. 2011;50(1):87–96.
Fu C, Jiang L, Hao S, Liu Z, Ding S, Zhang W, et al. Activation of the IL-4/STAT6 signaling pathway promotes lung cancer progression by increasing M2 myeloid cells. Front Immunol. 2019. https://doi.org/10.3389/fimmu.2019.02638.
Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380(6575):630–3.
Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4(3):313–9.
Wurster AL, Rodgers VL, White MF, Rothstein TL, Grusby MJ. Interleukin-4-mediated protection of primary B cells from apoptosis through Stat6-dependent up-regulation of Bcl-xL. J Biol Chem. 2002;277(30):27169–75.
Yevshin I, Sharipov R, Kolmykov S, Kondrakhin Y, Kolpakov F. GTRD: a database on gene transcription regulation-2019 update. Nucleic Acids Res. 2019;47(D1):D100–5.
McLeay RC, Bailey TL. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinform. 2010;11(1):165.
Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36.
Maier E, Wirnsberger G, Horejs-Hoeck J, Duschl A, Hebenstreit D. Identification of a distal tandem STAT6 element within the CCL17 locus. Hum Immunol. 2007;68(12):986–92.
Arthur SE, Jiang A, Grande BM, Alcaide M, Cojocaru R, Rushton CK, et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat Commun. 2018;9(1):4001.
Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256–65.
Dreval K, Hilton LK, Cruz M, Shaalan H, Ben-Neriah S, Boyle M, et al. Genetic subdivisions of follicular lymphoma defined by distinct coding and non-coding mutation patterns. Blood. 2023;142:561–73.
Häberle MT, Kelsch E, Dorsch K, Möller P, Ritz O. Knock-down of BCL6 / STAT6 sensitizes primary B cell lymphoma cells for treatment with current therapeutic agents. Oncoscience. 2014;1(4):283–6.
Baus D, Nonnenmacher F, Jankowski S, Döring C, Bräutigam C, Frank M, et al. STAT6 and STAT1 are essential antagonistic regulators of cell survival in classical Hodgkin lymphoma cell line. Leukemia. 2009;23(10):1885–93.
Tweeddale ME, Lim B, Jamal N, Robinson J, Zalcberg J, Lockwood G, et al. The presence of clonogenic cells in high-grade malignant lymphoma: a prognostic factor. Blood. 1987;69(5):1307–14.
Epstein AL, Levy R, Kim H, Henle W, Henle G, Kaplan HS. Biology of the human malignant lymphomas. IV. Functional characterization of ten diffuse histiocytic lymphoma cell lines. Cancer. 1978;42(5):2379–91.
Beckwith M, Longo DL, O’Connell CD, Moratz CM, Urba WJ. Phorbol ester-induced, cell-cycle-specific, growth inhibition of human B-lymphoma cell lines. JNCI J Natl Cancer Inst. 1990;82(6):501–9.
Seidel HM, Milocco LH, Lamb P, Darnell JE Jr, Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995;92(7):3041–5.
Chen HC, Reich NC. Live cell imaging reveals continuous STAT6 nuclear trafficking. J Immunol. 2010;185(1):64–70.
Gaydosik AM, Queen DS, Trager MH, Akilov OE, Geskin LJ, Fuschiotti P. Genome-wide transcriptome analysis of the STAT6-regulated genes in advanced-stage cutaneous T-cell lymphoma. Blood. 2020;136(15):1748–59.
Kapp U, Yeh W-C, Patterson B, Elia AJ, Kägi D, Ho A, et al. Interleukin 13 is secreted by and stimulates the growth of Hodgkin and Reed-Sternberg cells. J Exp Med. 1999;189(12):1939–46.
Mainou-Fowler T, Proctor SJ, Taylor PR. Interleukin 4 production by peripheral blood lymphocytes in patients with classical Hodgkin lymphoma. Leuk Res. 2004;28(2):159–66.
Newcom SR, Ansari AA, Gu L. Interleukin-4 is an autocrine growth factor secreted by the L-428 Reed-Sternberg cell. Blood. 1992;79(1):191–7.
Skinnider BF, Mak TW. The role of cytokines in classical Hodgkin lymphoma. Blood. 2002;99(12):4283–97.
Sugimoto T, Watanabe T. Follicular lymphoma: the role of the tumor microenvironment in prognosis. J Clin Exp Hematop. 2016;56(1):1–19.
Kotlov N, Bagaev A, Revuelta MV, Phillip JM, Cacciapuoti MT, Antysheva Z, et al. Clinical and biological subtypes of B-cell lymphoma revealed by microenvironmental signatures. Cancer Discov. 2021;11(6):1468–89.
Wirnsberger G, Hebenstreit D, Posselt G, Horejs-Hoeck J, Duschl A. IL-4 induces expression of TARC/CCL17 via two STAT6 binding sites. Eur J Immunol. 2006;36(7):1882–91.
Ye T, Zhang X, Dong Y, Liu J, Zhang W, Wu F, et al. Chemokine CCL17 affects local immune infiltration characteristics and early prognosis value of lung adenocarcinoma. Front Cell Dev Biol. 2022. https://doi.org/10.3389/fcell.2022.816927.
Mizukami Y, Kono K, Kawaguchi Y, Akaike H, Kamimura K, Sugai H, et al. CCL17 and CCL22 chemokines within tumor microenvironment are related to accumulation of Foxp3+ regulatory T cells in gastric cancer. Int J Cancer. 2008;122(10):2286–93.
Kanagawa N, Niwa M, Hatanaka Y, Tani Y, Nakagawa S, Fujita T, et al. CC-chemokine ligand 17 gene therapy induces tumor regression through augmentation of tumor-infiltrating immune cells in a murine model of preexisting CT26 colon carcinoma. Int J Cancer. 2007;121(9):2013–22.
Maeda S, Nakazawa M, Uchida M, Yoshitake R, Nakagawa T, Nishimura R, et al. Foxp3(+) regulatory T cells associated with CCL17/CCR4 expression in carcinomas of dogs. Vet Pathol. 2020;57(4):497–506.
Nakayama T, Hieshima K, Nagakubo D, Sato E, Nakayama M, Kawa K, et al. Selective induction of Th2-attracting chemokines CCL17 and CCL22 in human B cells by latent membrane protein 1 of Epstein–Barr virus. J Virol. 2004;78(4):1665–74.
Liu B, Lin Y, Yan J, Yao J, Liu D, Ma W, et al. Affinity-coupled CCL22 promotes positive selection in germinal centres. Nature. 2021;592(7852):133–7.
Cencini E, Fabbri A, Schiattone L, Sicuranza A, Mecacci B, Granai M, et al. Prognostic impact of tumor-associated macrophages, lymphocyte-to-monocyte and neutrophil-to-lymphocyte ratio in diffuse large B-cell lymphoma. Am J Blood Res. 2020;10(4):97–108.
Ciavarella S, Vegliante MC, Fabbri M, De Summa S, Melle F, Motta G, et al. Dissection of DLBCL microenvironment provides a gene expression-based predictor of survival applicable to formalin-fixed paraffin-embedded tissue. Ann Oncol. 2018;29(12):2363–70.
Locke FL, Ghobadi A, Jacobson CA, Miklos DB, Lekakis LJ, Oluwole OO, et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1–2 trial. Lancet Oncol. 2019;20(1):31–42.
Albanyan O, Chavez J, Munoz J. The role of CAR-T cell therapy as second line in diffuse large B-cell lymphoma. Ther Adv Hematol. 2022;13:20406207221141510.
Acknowledgements
This work was supported by operating grants from the Leukemia and Lymphoma Society of Canada/Societe de Leucemie & Lymphome Du Canada and Cancer Research Society (to K.K.M), the Canadian Cancer Society (to N.A.J) and a Cole Foundation Transition Grant (to S.V.dR). A.B. and M.J.A. were supported by Ph.D. Fellowships from the Cole Foundation. M.J.A was additionally supported by a Fonds de Recherche du Quebec Doctoral Training Fellowship. K.M.C. is a recipient of the Scholarship for the Next Generation of Scientists from the Cancer Research Society. R.D.M. was supported by Michael Smith Foundation for Health Research Scholar. The lymphoma biobank is supported by funds from the Jewish General Hospital foundation. The Genomic Variation in Diffuse Large B Cell Lymphomas study was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services. The datasets have been accessed through the NIH database for Genotypes and Phenotypes (dbGaP). We also acknowledge the ICGC MALY-DE project for providing access to their data. Aligned reads for those genomes were obtained through a DACO-approved project (to RDM) using a virtual instance on the Cancer Genome Collaboratory.
Author information
Authors and Affiliations
Contributions
All authors participated in research design and conceptualization. AB, MJA, SL, JK, RET, RB, NS, KM, CG, and RT conducted experiments. AB, MJA, SL, JK, RET, MS, KM, CG, MS, KOK, KMC, RDM, and SD performed data analyses. RS, RDM, SD, SA, and NAJ acquired patient samples. SVdR, NAJ, and KKM acquired funding. AB, MJA, and KKM wrote initial manuscript drafts. All authors approved of the final manuscript version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Benoit, A., Abraham, M.J., Li, S. et al. STAT6 mutations enriched at diffuse large B-cell lymphoma relapse reshape the tumor microenvironment. Int J Hematol 119, 275–290 (2024). https://doi.org/10.1007/s12185-023-03692-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-023-03692-x