Abstract
Phytoliths constitute microscopic SiO2-rich biominerals formed in the cellular system of many living plants and are often preserved in soils, sediments and artefacts. Their analysis contributes significantly to the identification and study of botanical remains in (paleo)ecological and archaeological contexts. Traditional identification and classification of phytoliths rely on human experience, and as such, an emerging challenge is to automatically classify them to enhance data homogeneity among researchers worldwide and facilitate reliable comparisons. In the present study, a deep artificial neural network (NN) is implemented under the objective to detect and classify phytoliths, extracted from modern wheat (Triticum spp.). The proposed methodology is able to recognise four phytolith morphotypes: (a) Stoma, (b) Rondel, (c) Papillate, and (d) Elongate dendritic. For the learning process, a dataset of phytolith photomicrographs was created and allocated to training, validation and testing data groups. Due to the limited size and low diversity of the dataset, an end-to-end encoder-decoder NN architecture is proposed, based on a pre-trained MobileNetV2, utilised for the encoder part and U-net, used for the segmentation stage. After the parameterisation, training and fine-tuning of the proposed architecture, it is capable to classify and localise the four classes of phytoliths in unknown images with high unbiased accuracy, exceeding 90%. The proposed methodology and corresponding dataset are quite promising for building up the capacity of phytolith classification within unfamiliar (geo)archaeological datasets, demonstrating remarkable potential towards automatic phytolith analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biomineralisation, the natural process by which living organisms produce mineral accumulations of diverse shapes/forms and sizes (Lowenstam 1981; Lowenstam and Weiner 1989), has been a vital process for terrestrial plants over their > 400 myr evolution history (Perry 2003; Trembath-Reichert et al. 2015). Biomineralisation in the plant kingdom may act as an agent for mechanical support, protection, detoxification, photosynthesis optimisation, and these functions may depend, among others, on the geometrical and morphological attributes of the plant biominerals (Meunier et al. 2017; Pierantoni et al. 2018; Strömberg et al. 2016). Phytoliths constitute microscopic SiO2-rich biominerals (commonly 10–30 μm in diameter and up to 200 μm in length), formed in the cellular system (within or between the cells) of many living plants (Bauer et al. 2011; Piperno 2006 and references therein; Shillito 2018). Phytoliths are often preserved (through decay in-situ mechanisms) in soils and sediments [and to a lesser extent on artefacts (e.g. Wendrich and Ryan 2012) or ecofacts (e.g. Cummings and Magennis 1997)] during the decomposition/burning of plant material, and their analysis contributes significantly to the identification and study of botanical remains in (paleo)ecological and archaeological contexts (Meunier and Collin 2001; Neumann et al. 2017; Rosen 1992), especially when used in conjunction with other archaeobotanical proxies (García-Granero et al. 2021; Horrocks 2005; Yao et al. 2012).
The different tissues in the plant organs have cells that often present distinct morphologies, hence, certain phytolith morphotypes relate to specific parts of the plant (e.g. inflorescence, leaves, stem) that may correlate with specific past (or present) human choices and activity that took place in the contexts where phytoliths are recovered (Esteban et al. 2018; Harvey and Fuller 2005; Tsartsidou et al. 2009). Furthermore, phytolith shape/form variation (e.g. rounded, angular) may be of high importance in discriminating between plant species (Ball and Brotherson 1992; Hošková et al. 2021; Twiss et al. 1969). In order to secure reliable information regarding phytolith assemblages, it is important that phytoliths remain morphologically and mineralogically/chemically unaltered at the highest possible level (Andriopoulou 2020; Cabanes and Shahack-Gross 2015; Jenkins 2009). Understanding the processes involved in the formation and preservation of phytoliths of defined shapes/forms and sizes remain challenging. Optical light microscopy (OLM), scanning electron microscopy (SEM), X-ray micro-computed tomography (X-ray MicroCT), confocal laser scanning microscopy (CLSM) and fluorescence microscopy (FL) constitute effective tools for phytolith examination and assessment of their geometry and morphology. The method of observation depends on the specific research questions (e.g. Andriopoulou et al. 2019; Fishkis et al. 2010; Gallaher et al. 2020; George et al. 2020; Puppe and Leue 2018; Russ and Rovner 1989; Soukup et al. 2014; Tsutsui et al. 2016).
Phytoliths are typically classified by their morphological characteristics, the plant taxon (or habitat type) and/or the anatomical plant part where they are formed onto (Madella et al. 2005; Neumann et al. 2019). Traditional phytolith classification (i.e. prediction of the class of an unknown sample) is mainly based on qualitative criteria and performed manually using OLM, which is usually time-consuming and may inherit human observer biases. Over the last few decades, there has been an increasing interest in the quantitative criteria using morphometrics (measurements of phytolith shape/form and/or size) based on semi-automatic image analysis (Ball and Brotherson 1992; Ball et al. 2016 and references therein). Computer-assisted approaches (geometric modern morphometrics) have been also conducted to obtain more complex morphometric parameters of individual (single-cells) or sometimes articulated phytoliths (multi-cells) (e.g. Ball et al. 2017; Hošková et al. 2021; Niu et al. 2021; Out et al. 2014) for the improvement of taxonomic resolution, especially when diagnostic features of phytoliths overlap. Despite the valuable advances in the phytolith research, methods, techniques and data collection vary considerably and thus the comparability of the results remains restricted.
An emerging challenge is the automatic classification of phytoliths that may enhance data homogeneity among researchers, reduce potential subjective observer bias and facilitate reliable comparisons, given that the method of microscopy remains the same. Evett and Cuthrell (2016) established the conceptual framework for a computer-assisted morphometric-based phytolith analysis and classification system (i.e. data acquisition, analysis, classification, integration, dissemination, accessibility). In recent years, interest has shifted towards the application of machine and deep learning algorithms on phytolith analysis. Deep learning (DL) is a new subfield of machine learning, which utilises complex (“deep”) artificial neural networks in order to learn higher level of data representations without the need of feature engineering, a time-consuming procedure in order to fine-tune the traditional machine learning algorithms (Goodfellow et al. 2016). The automatic phytolith classification approach has the advantage to classify morphotypes with a higher unbiased precision, and may be continuously refined, as more data become available.
Despite the significant contributions made in this direction, studies on phytolith classification using machine learning methodologies (Cai and Ge 2017; Diez-Pastor et al. 2020; Gallaher et al. 2020) remain extremely limited, while to the best of our knowledge, Andriopoulou et al. 2021 (additional data are given in the Online Resource 1) is the first study that utilised deep neural networks (deep learning) in (single- and multi-cell) phytolith classification (genus: Triticum). Recently, Berganzo-Besga et al. (2022) have also presented an algorithm for automatic classification of multi-cell phytoliths (genera: Avena, Hordeum and Triticum) with a high level of accuracy, using a convolutional neural network (CNN) architecture and three types of data augmentation techniques. Cai and Ge (2017) focused on the classification of short cell phytoliths extracted from several plant taxa. Among the four classification algorithms that they evaluated, support vector machines (SVM) and k-nearest neighbors (KNN) provided the highest performance in terms of accuracy. According to the comparison of six classification algorithms (with three different sets of features/attributes) applied on sediment phytoliths (composed of eight classes, i.e. morphotypes) by Díez-Pastor et al. (2020), the most accurate results were obtained using SVM (in accordance with Cai and Ge 2017) and random forests (RFs). Gallaher et al. (2020) explored the possibilities of three-dimensional (3D) geometric morphometrics of phytoliths (instead of the typical 2D), and through the study of fossil grass silica short cell phytoliths and the evaluation of six classification algorithms, they further developed an automatic approach to phytolith classification and analysis of shape evolution. Based on their results, linear discriminant analysis (LDA) consistently exhibited the highest accuracy with the exception of a case of logistic model trees (LMT), while SVM and multi-layer perceptron (MLP) were the lowest-performing among the tested algorithms.
In order to further the development of automatic phytolith classification, in the present study and based on our previous findings as shown in the Online Resource 1 (Andriopoulou et al. 2021), a deep artificial neural network (NN) with an encoder-decoder architecture is implemented to classify phytoliths extracted from modern wheat and digitally imaged using OLM. We propose an image-segmentation methodology, which is capable of detecting four phytolith morphotypes: (a) Stoma, (b) Rondel, (c) Papillate, and (d) Elongate dendritic. The resulting annotated dataset of phytolith photomicrographs was allocated to training, validation and testing data groups, feeding the deep neural network, in order to create a model that classifies and localises the four classes of phytoliths in the predicted images with high unbiased accuracy. The main contribution of this study is the implementation of a deep learning architecture capable to classify phytoliths efficiently using a limited size of phytolith dataset paving the way towards its application across different plant taxa and different methods of microscopy and phytolith representation.
Materials and methods
Phytolith extraction and image acquisition
Wheat (Triticum spp.) was selected because it produces large quantities of phytoliths (Albert and Weiner 2001), and its impact on economy since prehistory has been well documented (Zohary et al. 2012). Humans have been using wheat and its derivatives over time for food, beverages, medicine, animal fodder or straw for bedding and for other uses such as a structural material, fuel, temper for pottery/mudbricks and also for industrial purposes (Dzhanfezova 2020; Valamoti 2009 and references therein). Wheat is not only used in everyday life settings but also in feasts and rituals (Megas 2001; Miligou-Markantoni 2006). The last decade there has been a growing interest in the identification of cereals or cereal preparations (Fuller and Gonzalez Carretero 2018; Liu et al. 2020; Valamoti 2011), and especially wheat (often when its chaff has been processed) in (geo)archaeological contexts using, among others, phytolith analysis (Ball et al. 1999; Berlin et al. 2003; Cabanes et al. 2012).
Phytoliths were extracted from Triticum aestivum, Triticum dicoccum, Triticum durum and Triticum spelta originated from ecological crops (four plant plots) in Lleida, Catalonia, Spain, where extensive experimental archaeobotanical research is conducted (Alonso et al. 2013; Andriopoulou 2014 [unpublished Master’s thesis]; Asensio et al. 2011). In order to further understand changes in the phytolith assemblage that are related to the particular stage in the life cycle of the plant, phytoliths were also extracted using the same laboratory method from Triticum durum and Triticum monococcum (two plant plots) that we ecologically cultivated in an experimental field (Fig. 1) under similar growing conditions at Chania, Crete, Greece (Andriopoulou 2020).
Harvesting two wheat plots in the experimental field at Chania, Crete, Greece. Soil and control samples for future phytolith analyses were also taken from the plots at different depths, before and after the cultivation period [source: Andriopoulou 2020 (photographs: Michelakis and Andriopoulou)]
The phytoliths were extracted from dried mature plants using a slightly modified version of the dry method (Parr et al. 2001). Andriopoulou and Christidis (2020, 2021) have focused on the impact of the two conventional phytolith extraction methods on the preservation status of the phytoliths, and suggested that dry method has certain advantages compared with the wet method. The entire plants including leaves, stems and inflorescences were manually cut, stored in bakers and washed twice in deionised water using the sonicator bath for ca. 15 min, to remove impurities attached to the plant surface. The root system, when available, was separated from the rest of the plant material and stored, to prevent soil contamination. The washed plant material was dried overnight in an oven at ~ 120 °C and weighted. The dried parts of the plant material were transferred to crucibles and burned in a muffle furnace at ~ 550 °C for 4 h.
The resulting ashes were transferred to 50 mL falcon tubes. Ten (10) mililitre of 10% hydrochloric acid (HCl) was added to the tubes to dissolve the carbonate minerals, with extra HCl being added when necessary, until bubbling ceased. The samples were then vortexed for a few seconds and centrifuged at 3000 rpm for ca. 5 min. The supernatant was discarded and 10 mL of deionised H2O was added to the tubes, which were centrifuged under the same conditions as before. Washing with H2O was repeated twice and the samples were dried overnight at ~ 80 °C. Subsequently, the samples were transferred to watch glasses and 30 mL of 33% hydrogen peroxide (H2O2) was added, to eliminate the remaining organic material. The watch glasses were heated on a hotplate at ~ 80 °C to accelerate oxidation. Further H2O2 was gradually added until no charcoal/organic remains were macroscopically observed. The remaining peroxide was eliminated by centrifugation at 3000 rpm for ca. 5 min followed by washing with deionised H2O. The samples were dried overnight at ~ 80 °C, softly scratched off from the watch glasses with a spatula and weighted.
The remaining fraction after the elimination of carbonates and organic material is named Acid Insoluble Fraction (AIF) (Albert and Weiner 2001). Three different slides were prepared from each sample using 0.5–0.9 mg of the AIF that is composed mainly of phytoliths. The mounting medium used was Entellan (Merck). Photomicrographs of phytoliths were acquired using OLM (an Olympus BX-41 and a Zeiss Axioscop 40 microscope with an attached digital camera; using the Delta Pix Viewer and the Leica Application Suite software). Phytolith morphotypes with clear morphological outer borders (outline) and morphologically unaltered at the highest possible level were identified based on the standard literature (Madella et al. 2005; Neumann et al. 2019; Piperno 2006; Twiss et al. 1969; Zurro 2018).
Dataset description
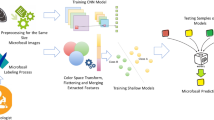
In the next step, the phytolith photomicrographs were further processed in order to feed the neural network (NN) model. It is worth pointing out that the qualitative dataset creation is crucial for the learning process of neural network algorithms. Initially, the photomicrographs are manually annotated (establishing its contours and colorizing with a different colour per class) forming four classes of phytoliths linked to different anatomical plant parts (i.e. leaves, stem, and inflorescence). The resulting dataset, which includes the raw images (photomicrographs) and the corresponding annotated images (ground truth in deep learning terminology), describes the aforementioned phytolith classes: (a) Stoma, (b) Rondel, (c) Papillate, and (d) Elongate dendritic.
Subsequently, the dataset (images/annotations) was split in training, validation and testing sets with a percentage of 70-20-10% respectively, feeding a deep NN with the aim of learning and subsequently detecting and localising the four classes of phytoliths using semantic-segmentation (Wang et al. 2022). The resulting size of dataset was 250 pairs of images, which is quite small for efficient learning processes, while it was one of the main issues that the methodology had to confront.
Neural network architecture
In Andriopoulou et al. (2021), which was the initial approach of the present study, a fully convolutional neural network (FCN) was implemented and trained from scratch using the aforementioned dataset in order to detect and segment the phytolith morphotypes automatically (Online Resource 1). More specifically, a U-net based architecture was implemented because of its high efficiency in limited size datasets (Ronneberger et al. 2015). The U-shaped model of U-net was divided in two main parts: (a) the encoder, which downscales the images, increasing the feature maps and learns from the content of the training dataset, and (b) the decoder, which upscales the images, decreasing the feature maps and conducts precise localisation of the detected phytoliths. Simultaneously, four connections (skip connections) among different levels of the encoder, transfer information to the decoder in order to maintain the spatial details of images with the aim to reconstruct them.
Although the accuracy of the above approach delivered positive results, the limited size of the dataset in combination with the multi-class classification (which injects further complexity to the process) and the use of higher content complexity images with low diversity for a learning procedure, prevented the methodology from achieving more accurate results.
In the present study, an end-to-end encoder-decoder NN architecture was implemented based on U-net (Ronneberger et al. 2015) and MobileNetV2 (Sandler et al. 2018) neural networks. More specifically, a pre-trained MobileNetV2 was utilised for the encoder part, while the decoder of U-net was used for the segmentation stage, maintaining the skip connections that U-net implements between the encoder and decoder. The basic idea of MobileNetV2 architecture is the depth-wise separable convolutions, a DL technique, which reduces the network parameters and the required computational power. The depth-wise separable convolutions are the core components that compose the two main blocks of MobileNetV2 architecture called inverted residual blocks (Fig. 2) (Sandler et al. 2018).
In the present methodology, the MobileNetV2 is pre-trained with ImageNet (Deng et al. 2009), a well-known image dataset with more than 14 million of general purpose photographs and more than 1 million groups of classes. Thus, during the training of the proposed NN, the MobileNetV2 transfers information to the decoder which is related with the abstract characteristics of the ImageNet dataset (shapes, texture, patterns etc.), facilitating and accelerating the learning process for phytolith classification and segmentation. The process above is called transfer learning in deep learning terminology.
After the training process, the proposed architecture was able to classify and localise the four phytolith morphotypes (Stoma, Rondel, Papillate, and Elongate dendritic) in photomicrographs completely unknown for the implemented NN. Figure 3 presents the end-to-end architecture of the proposed NN.
Proposed NN architecture for phytolith classification and segmentation. The input image (photomicrograph), which is an RGB image with 256 × 256 pixel resolution feeds the encoder, the pre-trained with ImageNet weights MobileNetV2. In the next step, the decoder from U-net model proceeds to the segmentation stage, while at the end the Softmax function is used as the last activation function of the NN and outputs the predicted image
Results
During experimentation, the final parameters of the proposed architecture were defined, by fine-tuning the related network parameters and optimising its efficiency. Concerning the loss function, the Tversky loss function was utilised, which is a quite effective loss function for unbalanced datasets (Salehi et al. 2017). Regarding the hyper parameters of the model, the Nadam optimiser was used (Dozat 2016), the learning rate was set in 0.0001 while the learning process completed in 125 epochs with a batch size equal to 6. The performance of the proposed architecture was assessed during training and validation, by calculating three well-known metrics for semantic-segmentation: dice- coefficient, recall and precision (Seo et al. 2021). The NN architecture was developed using the KerasFootnote 1/TensorflowFootnote 2 framework while for the training process the Google colab platformFootnote 3 with Tesla K80 GPU was utilised.
The results demonstrate that the model classifies and localises the phytolith morphotypes (Stoma, Rondel, Papillate, and Elongate dendritic) in the predicted images with high level of accuracy. Table 1 presents the metrics results of the proposed NN after the end of the training process and Fig. 4 depicts the learning curves of the loss function in training and validation data.
As presented in Table 1, all metrics in testing and validation data were greater than 90%, while as observed in Fig. 4, both learning curves conclude in a loss function value of about 0.08, which denotes a high level of accuracy. It is worth pointing out that all the metric values between training and validation were similar in each metric, which means that the proposed NN does not overfit. In other words, the present architecture performs with high accuracy in validation data during the learning process, showing that it does not memorise the training data (overfitting), instead it produces valid predictions.
Beyond the evaluation during the training process, the proposed NN was further tested aiming to evaluate the classification and localisation performance for each specific phytolith class, using the testing data, which constitutes 10% of the whole dataset.
More specifically, the overall accuracy of the proposed NN is 0.92 while the Precision, Recall, Dice-coefficient and Jaccard score for each class are presented in Table 2:
As presented in Table 2, the Precision, Recall and Dice-coefficient of all the classes is about 0.60 or above while the Jaccard score is in a level of 0.40. The class “Stoma” provides the highest performance both localisation and classification while the class “Rondel” although achieves high classfication performance, it provides lower efficiency in the localisation since Jaccard score is in a level of 0.40. The classes “Elongate dendritic” and “Papillate” achieve similar performance with Dice-coefficient in a level of 0.50 and Jaccard score about 0.35.
The results are considered satisfactory taking into account the limited size of dataset, while in most images the phytoliths over quite small region in the image, a fact that increases the difficulty of accurate segmentation. It is worth noting that the background, which technically is considered as a class, is not included in the Table 2 because it would fictitiously increase the mean of the results since its performance is about 0.95 in all the metrics.
Visual results of the predicted images are present in Fig. 5. The phytolith photomicrographs from the testing data, which are data that the model has never seen before are depicted in the first column. The annotated images are present in the second column, while the predicted images are visualised in the third column.
Phytolith photomicrographs that the model has never seen before (1st column), annotated images (2nd column) and predicted images (3rd column). Regarding the colorisation, the class “Stoma” is represented with green colour, the class “Rondel” with red colour, the class “Papillate” with yellow colour and the class “Elongate dendritic” with purple colour
The proposed architecture produces robust and accurate results in phytolith photomicrographs with low image content complexity as present in the first three images (a-c) of Fig. 5. On the other hand, in higher content complexity images, although the accuracy of the present methodology is satisfactory, detecting all the types of classes, yet, the localisation of phytolith morphotypes is not as accurate as in a-c examples. For instance, in Fig. 5d, the one of two Papillate morphotypes are localised, while in Fig. 5f the model does not predict the class “Papillate” with high performance in terms of segmentation. On the other handin Fig. 5g, the proposed NN has predicted the class “Papillate” with high level of accuracy, while it has not detected the one of two Elongate dendritic morphotypes. However, in Fig. 5e the proposed NN achieves high performance in detection and localisation. It is worth mentioning that although it detects one more Rondel than the ground truth, indeed in this specific location, there is a feature that resembles a Rondel morphotype.
Discussion
Αs the experimentation proved, the present methodology achieved accurate classification and detection of four phytolith classes, while it encountered the aforementioned issue of a limited and unbalanced dataset. Initially, Tversky function was utilised as the loss function of the proposed NN, because of its efficiency in datasets with an unequal number of examples per class. Subsequently, data augmentation was used, a technique that distorts the images of dataset through rotation, translation, re-scaling, etc. in order to pseudo-increase the size of dataset and enrich its content. While these techniques improved the results, transfer learning encountered efficiently the dataset size issue. The proposed encoder-decoder NN architecture utilises a pre-trained MobileNetV2 NN as the encoder, while the U-net decoder is used to reconstruct and segment the photomicrographs. Thus, the present architecture is not trained from scratch; instead, it already holds an “experience” from millions of general purpose images.
Compared with the initial approach of this methodology (Andriopoulou et al. 2021), the proposed architecture extracts more accurate and robust results (Table 3; Fig. 6), while the required time of the learning process is significantly reduced. In Andriopoulou et al. 2021 (Online Resource 1), the U-net model used was trained from scratch with 250 phytolith images (training dataset), while the present architecture is trained with the same phytolith dataset utilising the knowledge of ImageNet dataset through the pre-trained MobileNetV2.
More specifically, to quantify the initial approach’s performance in terms of classification and segmentation, the metrics Precision, Recall, Dice-coefficient and Jaccard score were used and as presented in Table 3:
As presented in Table 3, the performance of the U-net model is decreased compared with the proposed NN. Concerning the classification performance of the two models, the initial model provides about two timesless performance, since U-net model’s Precision and Recall are in a level of 0.30 instead of the proposed NN, which are in a level of 0.65. Moreover, regarding the segmentation performance, the initial model provides about three times less efficiency, since U-net model’s Dice-coefficient and Jaccard score are in a level of 0.20 and 0.15 compared with the proposed NN, which are in a level of 0.65 and 0.40 respectively. However, the main drawback of the initial methodology is that the initial model provides quite low performance in “Papillate” class, as it is presented in Table 3.
Beyond the quantitative results, a sample of visual results for the initial and proposed architectures are presented in Fig. 6:
a-e Comparison results between the initial approach (Andriopoulou et al. 2021) and proposed architecture. In each example (a-e), the third column is the result from the proposed architecture, while the forth column is the corresponding result from the initial approach of this methodology (Online Resource 1). The first and second column is the raw and the ground truth image respecively. Regarding the colorisation, the class “Stoma” is represented with green colour, the class “Rondel” with red colour, the class “Papillate” with yellow colour and the class “Elongate dendritic” with purple colour
As depicted in Fig. 6, in images with uncomplicated content both architectures extract similar results (Fig. 6a, b), while in images with medium or high level of complexity (Fig. 6c, d, e), the performance of the initial approach is decreased. As depicted in Fig. 6b, while the proposed NN produces an accurate result, the U-net implementation of the initial approach detects the “Stoma” class but falsely localises more than one Rondel morphotypes while in Fig. 6c although the proposed NN provides satisfactory performance, the initial model does not detect all the “Stoma” features and localises many more Rondel morphotypes than the reference image. Moreover, in the Fig. 6d, the U-net model localises most of the Papillate morphotypes but classifies them as Rondel morphotypes, while it does not detect the “Elongate dendritic” class. Finally, in Fig. 6e while the proposed NN localises and classifies both classes, the initial model produces a quite inaccurate prediction.
Although the proposed NN provides satisfactory performance, one drawback is the limited segmentation accuracy in the classes “Elongated dendritic” and “Papillate” as presented in Fig. 5, More specifically, it does not accurately predict the ridges of the Elongate dendritic and Papillate morphotypes but it depicts the corresponding features with a more rounded and spherical shape. This issue is quantified by the segmentation metrics, since the model provides Dice-coefficient in a level of 0.55 and Jaccard score in a level of 0.35 in both classes which are decreased compared with the classes “Rondel” and “Stoma”. This shortcoming of the model could be οvercome, using a larger dataset.
However, it is clear that although the “experience” that was transfered in the proposed NN through the transfer learning technique, was derived from general purposed images, it offered an essential advantage since the fine-tuning of the final NN parameters was improved and accelerated, achieving higher level of effectiveness at the same time.
It is worth noting that the proposed NN, is able to detect and localise the four morphotypes among articulated (multi-cells) phytoliths, such as in the Fig. 5e, with respectable accuracy which is significantly complex for machine learning algorithms to achieve (e.g. Cai and Ge 2017). This is quite important, because it indicates that deep learning provides an added value in automatic phytolith classification. Most of the studies consider that deep learning requires thousands of data for an automatic classification (Díez-Pastor et al. 2020), while the present study proves that only 250 pairs of phytolith images are enough to build a deep learning model for phytolith classification.
Although DL automates all the human effort that is needed to fine-tune machine learning algorithms, which is called feature engineering (Goodfellow et al. 2016), supervised classification requires the time-consuming process of manual image annotation in order to build the dataset for the training and validation process. The dataset creation with raw and annotated images is one of the main issues of deep learning community in each implementation. For this reason, huge projects for dataset creation and development are being developed either for general or specific purposes. In the same direction, phytolith community could create a collective and dynamic platform for data creation and annotation in order to reinforce and accelerate the next phytolith classification methodologies. The phytolith dataset existence is crucial because it automates the most time-consuming part of a DL implementation, while it facilitates and encourages the development of more NN models for phytolith classification. As a result, phytolith researchers could integrate a trained NN in their methodology loading just one file, building a ready-to-use automatic phytolith classification system.
The present study is a quite promising methodology for automatic phytolith classification through semantic-segmentation with remarkable potential for further development. The number of phytolith morphotypes to be detected could be increased, adding more classes with multiple spatial orientations for even more useful and robust results. The application of DL algorithms on phytolith analysis may further include the sub-classification of phytoliths subjective or based on more detailed geometrical and/or morphological features/attributes, combining insights from 3D imaging results.
The proposed methodology may find future applications in the study of other (archaeo)botanical microremains, such as pollen and starch granules that are often used in conjuction with phytolith analysis (e.g. Cummings and Magennis 1997; Holst et al. 2007; Yao et al. 2012). Previous studies have already combined human input with computer vision and employed datasets for automatic pollen (e.g. Dunker et al. 2021; Gonçalves et al. 2016; Kubera et al. 2021) or starch granules classification (e.g. Arráiz et al. 2016; Earles et al. 2018; Wilson et al. 2010).
Our long-term goal is to facilitate the establishment of automatic phytolith analysis and expand the archaeobotanical network based on three main contributions:
-
Free available code under an open source license (e.g. GPL3Footnote 4) for a deep learning phytolith classification system where every researcher could use, modify and improve.
-
Already trained NNs for phytolith classification, where non-DL experts could integrate just one file in their methodology with the earned “experience” of the NN and the model would be ready to classify without all the above procedure.
-
A rich, free accessible database with a plethora of annotated phytolith images, which would be a valuable source for new deep learning implementations in phytolith classification.
Conclusions
An implementation of an end-to-end encoder-decoder NN architecture for phytolith classification and segmentation is proposed. The present architecture is based on MobileNetV2 and U-net, while transfer learning is utilised through the MobileNetV2 in order to improve the NN efficiency and encounter the limited dataset issue. The methodology is able to detect four phytolith classes (Stoma, Rondel, Papillate, and Elongate dendritic) in photomicrographs (using OLM) completely unknown for the NN and achieve robust and accurate results. Although additional training samples and plant species datasets are required to optimise the results, the present dataset extracted from modern plant material is promising for building up the capacity of phytolith classification within unfamiliar (geo)archaeological datasets for the study of the diachronic human (or non-human animal) and plant interactions.
Data availability
The dataset of the current study is publicly available in the Kaggle platform: https://www.kaggle.com/datasets/georgepetrakis/phytolith-photomicrographs.
References
Albert RM, Weiner S (2001) Study of phytoliths in prehistoric ash layers using a quantitative approach. In: Meunier JD, Coline F (eds) Phytoliths: applications in Earth Sciences and Human History. AA Balkema Publishers, Lisse, pp 251–266
Alonso N, Antolín F, López D, Cantero FJ, Prats G (2013) The effect of dehusking on cereals: experimentation for archaeobotanical comparison. In: Anderson PC, Cheval C, Durand A (eds) Regards croisés sur les outilsliés au travail des végétaux [An interdisciplinary focus on plant-working tools]. APDCA, Antibes, pp 131–144
Andriopoulou NC (2014) Phytolith analysis of cereals, modern soils and experimental crop proccessing activities: application to archaeology. Unpublished Master's thesis, International Erasmus Mundus Master in Quaternary and Prehistory
Andriopoulou NC, Michailidis D, Christidis GE, Partsinevelos P (2019) 3D representation of biominerals integrating microscopy and photogrammetry: implications in geoarchaeology. Geophysical Research Abstracts vol. 21, EGU2019-10780-1. European Geoscience Union General Assembly, 7–12 April 2019, Vienna, Austria
Andriopoulou NC, Christidis GE (2020) Multi-analytical characterisation of wheat biominerals: impact of methods of extraction on the mineralogy and chemistry of phytoliths. Archaeol Anthropol Sci 12(186). https://doi.org/10.1007/s12520-020-01091-5
Andriopoulou NC (2020) Biomineralisation and biogeochemical mobility of chemical elements: phytolith dissolution and its implication in geoarchaeology. Doctoral thesis, School of Mineral Resources Engineering, Technical University of Crete, Greece. https://phdtheses.ekt.gr/eadd/handle/10442/48192. Accessed 22 Nov 2020
Andriopoulou NC, Christidis GE (2021) Preservation of biominerals: impact of laboratory extraction methods on the physicochemical characteristics of SiO2-rich phytoliths, XXXV Panhellenic Conference on Solid State Physics and Materials Science (virtual), 26–29 Sept 2021, Athens, Greece (Congress Center of NCSR “Demokritos”)
Andriopoulou NC, Petrakis G, Partsinevelos P (2021) Phytolith classification using deep learning: implementation of a U-net neural network for morphotype image-segmentation, International Conference (virtual), 12th International Meeting for Phytolith Research, 6–11 Sept 2021, Kiel, Germany, in the frame of the 27th annual meeting of the European Association of Archaeologists
Arráiz H, Barbarin N, Pasturel M, Beaufort L, Domínguez-Rodrigo M, Barboni D (2016) Starch granules identification and automatic classification based on an extended set of morphometric and optical measurements. J Archaeol Sci Rep 7:169–179. https://doi.org/10.1016/j.jasrep.2016.03.039
Asensio D, Cardona R, Colell I, García Dalmau C, Ferrer C, Morer de Llorens J, Saula O, Pou J (2011) El fossat del poblat ibèric dels Estinclells (Verdú, l’Urgell), segle III aC. [The moat of the Iberian village Estinclells (Verdu, Urgell), 3rd entury BC] evista d’ Arquelogia de Ponent 21:121–128 (in Catalan). https://raco.cat/index.php/RAP/article/view/252322
Ball TB, Brotherson JD (1992) The effect of varying environmental conditions on phytolith morphometries in two species of grass (Bouteloua curtipendula and Panicum virgatum). Scanning Microsc 6:1163–1181. https://digitalcommons.usu.edu/microscopy/vol6/iss4/27
Ball TB, Gardner JS, Anderson N (1999) Identifying inflorescence phytoliths from selected species of wheat (Triticum monococcum, T. dicoccon, T. dicoccoides and T. aestivum) and barley (Hordeum vulgare and H. spontaneum) (Gramineae). Am J Bot 86:1615–1623. https://doi.org/10.2307/2656798
Ball TB, Davis AL, Evett RR, Ladwig JL, Tromp M, Out WA, Portillo M (2016) Morphometric analysis of phytoliths: recommendations towards standardization from the International Committee for Phytolith Morphometrics. J Archaeol Sci 68:106–111. https://doi.org/10.1016/j.jas.2015.03.023
Ball TB, Vrydaghs L, Mercer T, Pearce M, Snyder S, Lisztes-Szabó Z, Pető A (2017) A morphometric study of variance in articulated dendritic phytolith wave lobes within selected species of Triticeae and Aveneae. Veg Hist Archaeobot 26:85–97. https://doi.org/10.1007/s00334-015-0551-x
Bauer P, Elbaum R, Weiss IM (2011) Calcium and silicon mineralization in land plants: transport, structure and function. Plant Sci 180:746–756. https://doi.org/10.1016/j.plantsci.2011.01.019
Berganzo-Besga I, Orengo HA, Lumbreras F, Paloma A, Ramsey MN (2022) Automated detection and classification of multi-cell phytoliths using deep learning-based algorithms. J Archaeol Sci 148:105654. https://doi.org/10.1016/j.jas.2022.105654
Berlin AM, Ball TB, Thompson R, Herbert SC (2003) Ptolemaic agriculture, “Syrian wheat” and Triticum aestivum J Archaeol Sci 30:115–121. https://doi.org/10.1006/jasc.2002.0812
Cabanes D, Gadot Y, Cabanes M, Finkelstein I, Weiner S, Shahack-Gross R (2012) Human impact around settlement sites: a phytolith and mineralogical study for assessing site boundaries, phytolith preservation, and implications for spatial reconstructions using plant remains. J Archaeol Sci 3:2697–2705. https://doi.org/10.1016/j.jas.2012.04.008
Cabanes D, Shahack-Gross R (2015) Understanding fossil phytolith preservation: the role of partial dissolution in paleoecology and archaeology. PLoS ONE 10:e0125532. https://doi.org/10.1371/journal.pone.0125532
Cai Z, Ge S (2017) Machine learning algorithms improve the power of phytolith analysis: a case study of the tribe Oryzeae (Poaceae). J Syst Evol 55:377–384. https://doi.org/10.1111/jse.12258
Cummings LS, Magennis A (1997) A phytolith and starch record of food and grit in Mayan human tooth tartar. In: Pinilla A, Juan-Tresserras J, Machado MJ (eds) Estado actual de los estudios de fitolitos en suelos y plantas [The state-of-the-art of phytoliths in soils and plants], Monografias del Centro de Ciencias Medioambientales, Madrid, pp 211–218. https://doi.org/10.6067/XCV8V987PC
Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L (2009) Imagenet: A large-scale hierarchical image database. The IEEE conference on Computer Vision and Pattern Recognition, Miami, USA, 20–25 June 2009, pp 248–255
Díez-Pastor JF, Latorre-Carmona P, Arnaiz-González A, Ruiz-Pérez J, Zurro D (2020) “You are not my type”: an evaluation of classification methods for automatic phytolith identification. Microsc Microanal 26:1158–1167. https://doi.org/10.1017/S1431927620024629
Dozat T (2016) Incorporating Nesterov Momentum into Adam. ICLR Workshop 1:2013–2016
Dunker S, Motivans E, Rakosy D, Boho D, Mäder P, Hornick T, Knight TM (2021) Pollen analysis using multispectral imaging flow cytometry and deep learning. New Phytol 229:593–606. https://doi.org/10.1111/nph.16882
Dzhanfezova T (2020) Organic temper and the Early Neolithic pottery production: interpretational challenges. Acta Archaeol 91:61–87. https://doi.org/10.1111/j.1600-0390.2020.12228.x
Earles JM, Knipfer T, Tixier A, Orozco J, Reyes C, Zwieniecki MA, Brodersen CR, McElrone AJ (2018) In vivo quantification of plant starch reserves at micrometer resolution using X-ray microCT imaging and machine learning. New Phytol 218:1260–1269. https://doi.org/10.1111/nph.15068
Esteban I, Marean CW, Fisher EC, Karkanas P, Cabanes D, Albert RM (2018) Phytoliths as an indicator of early modern humans plant gathering strategies, fire fuel and site occupation intensity during the Middle Stone Age at Pinnacle Point 5–6 (South Coast, South Africa). PLoS ONE 13:e0198558. https://doi.org/10.1371/journal.pone.0198558
Evett RR, Cuthrell RQ (2016) A conceptual framework for a computer-assisted morphometric-based phytolith analysis and classification system. J Archaeol Sci 68:62–69. https://doi.org/10.1016/j.jas.2015.09.003
Fishkis O, Ingwersen J, Lamers M, Denysenko D, Streck T (2010) Phytolith transport in soil: a field study using fluorescent labeling. Geoderma 157:27–36. https://doi.org/10.1016/j.geoderma.2010.03.012
Fuller DQ, Gonzalez Carretero L (2018) The archaeology of Neolithic cooking traditions: archaeobotanical approaches to baking, boiling and fermenting. Archaeol Int 21:109–121. https://doi.org/10.5334/ai-391
Gallaher TJ, Akbar SZ, Klahs PC, Marvet CR, Senske AM, Clark LG, Strömberg CAE (2020) 3D shape analysis of grass silica short cell phytoliths: a new method for fossil classification and analysis of shape evolution. New Phytol 228:376–392. https://doi.org/10.1111/nph.16677
García-Granero JJ, Hatzaki E, Tsafou E, Ayala G, Serpetsidaki I, Bogaard A (2021) From storage to disposal: a holistic microbotanical approach to domestic plant preparation and consumption activities in Late Minoan Gypsades, Crete. J Archaeol Method Theory 28:307–331. https://doi.org/10.1007/s10816-020-09456-9
George N, Antony A, Ramachandran T, Hamed F, Kamal-Eldin A (2020) Microscopic investigations of silicification and lignification suggest their coexistence in tracheary phytoliths in date fruits (Phoenix dactylifera L). Front Plant Sci 11:977. https://doi.org/10.3389/fpls.2020.00977
Gonçalves AB, Souza JS, Da Silva GG, Cereda MP, Pott A, Naka MH, Pistor H (2016) Feature extraction and machine learning for the classification of brazilian Savannah pollen grains. PLoS ONE 11:e0157044. https://doi.org/10.1371/journal.pone.0157044
Goodfellow IJ, Bengio Y, Courville A (2016) Deep learning. The MIT Press, Cambridge
Harvey EL, Fuller DQ (2005) Investigating crop processing using phytolith analysis: the example of rice and millets. J Archaeol Sci 32:739–752. https://doi.org/10.1016/j.jas.2004.12.010
Holst I, Moreno J, Piperno D (2007) Identification of teosinte, maize and Tripsacum in Mesoamerica by using pollen, starch grains and phytoliths. Proc Natl Acad Sci 104:17608–17613. https://doi.org/10.1073/pnas.0708736104
Horrocks M (2005) A combined procedure for recovering phytoliths and starch residues from soils, sedimentary deposits and similar materials. J Archaeol Sci 32:1169–1175. https://doi.org/10.1016/j.jas.2005.02.014
Hošková K, Pokorná A, Neustupa J, Pokorný P (2021) Inter- and intraspecific variation in grass phytolith shape and size: a geometric morphometrics perspective. Ann Bot 127:191–201. https://doi.org/10.1093/aob/mcaa102
Jenkins E (2009) Phytolith taphonomy: a comparison of dry ashing and acid extraction on the breakdown of conjoined phytoliths formed in Triticum durum J Archaeol Sci 36:2402–2407. https://doi.org/10.1016/j.jas.2009.06.028
Kubera E, Kubik-Komar A, Piotrowska-Weryszko K, Skrzypiec M (2021) Deep learning methods for improving pollen monitoring. Sensors 21:3526. https://doi.org/10.3390/s21103526
Liu L, Li Y, Hou J (2020) Making beer with malted cereals and qu starter in the Neolithic Yangshao culture, China. J Archaeol Sci Rep 29:102134. https://doi.org/10.1016/j.jasrep.2019.102134
Lowenstam HA (1981) Minerals formed by organisms. Science 211:1126–1131. https://doi.org/10.1126/science.7008198
Lowenstam HA, Weiner S (1989) On biomineralization. Oxford University Press, New York
Madella M, Alexandre A, Ball T (2005) International Code for Phytolith Nomenclature 1.0. Ann Bot 96:253–260. https://doi.org/10.1093/aob/mci172
Megas G (2001) Ελληνικές γιορτές και έθιµα της λαϊκής λατρείας [Greek feasts and customs of folk worship]. Βιβλιοπωλείον της Εστίας [Hestia Bookstore], p 34 (in Greek)
Miligou-Markantoni M (2006) ∆ένδρα, φυτά, άνθη στον λαϊκό πολιτισµό των νεώτερων Ελλήνων [Trees, plants and flowers of folk culture of modern Greeks]. Ιδιωτική Έκδοση [Private Edition], p 663 (in Greek)
Meunier JD, Collin F (2001) Phytoliths - Applications in Earth Sciences and human history, 1st edn. AA Balkema Publishers, Lisse France
Meunier JD, Barboni D, Anwar-ul-Haq M, Levard C, Chaurand P, Vidal V, Grauby O, Huc R, Laffont-Schwob I, Rabier J, Keller C (2017) Effect of phytoliths for mitigating water stress in durum wheat. New Phytol 215:229–239. https://doi.org/10.1111/nph.14554
Neumann K, Chevalier A, Vrydaghs L (2017) Phytoliths in archaeology: recent advances. Veg Hist Archaeobot 26:1–3. https://doi.org/10.1007/s00334-016-0598-3
Neumann K, Strömberg CAE, Ball T, Albert RM, Vrydaghs L, Cummings LS, ICPT, International Committee for Phytolith Taxonomy ICPT (2019) International Code for Phytolith nomenclature (ICPN) 2.0. Ann Bot 124:189–199. https://doi.org/10.1093/aob/mcz064
Niu H, Shi J, Jie D (2021) Computer-assisted classification and discrimination of common phytoliths in the surface soils of different vegetation types in Northeast China. Rev Palaeobot Palynol 284:104326. https://doi.org/10.1016/j.revpalbo.2020.104326
Out WA, Grau JFP, Madella M (2014) A new method for morphometric analysis of opal phytoliths from plants. Microsc Microanal 20:1876–1887. https://doi.org/10.1017/S1431927614013270
Parr JF, Lentfer CJ, Boyd WE (2001) A comparative analysis of wet and dry ashing techniques for the extraction of phytoliths from plant material. J Archaeol Sci 28:875–886. https://doi.org/10.1006/jasc.2000.0623
Perry CC (2003) Silicification: the processes by which organisms capture and mineralize silica. Rev Mineral Geochem 54:291–327. https://doi.org/10.2113/0540291
Pierantoni M, Tenne R, Rephael B, Brumfeld V, van Casteren A, Kupczik K, Oron D, Addadi L, Weiner S (2018) Mineral deposits in Ficus leaves: morphologies and locations in relation to function. Plant Physiol 176:1751–1763. https://doi.org/10.1104/pp.17.01516
Piperno D (2006) Phytoliths. A comprehensive guide for archaeologists and paleoecologists. AltaMira Press, Lanham
Puppe D, Leue M (2018) Physicochemical surface properties of different biogenic silicon structures: results from spectroscopic and microscopic analyses of protistic and phytogenic silica. Geoderma 330:212–220. https://doi.org/10.1016/j.geoderma.2018.06.001
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, FrangI A (eds) Medical image computing and computer-assisted intervention. MICCAI 2015, vol 9351. Springer, Cham. https://doi.org/10.1007/978-3-319-24574-4_28
Rosen AM (1992) Preliminary identification of silica skeletons from Near Eastern archaeological sites: an anatomical approach. In: Rapp G, Mulholland SC (eds) Phytolith systematics. Advances in Archaeological and Museum Science, vol. 1. Springer, Boston, pp 129–147. https://doi.org/10.1007/978-1-4899-1155-1_7
Russ JC, Rovner I (1989) Stereological identification of opal phytolith populations from wild and cultivated Zea. Am Antiq 54:784–792. https://doi.org/10.2307/280682
Salehi SSM, Erdogmus D, Gholipour A (2017) Tversky loss function for image segmentation using 3D fully convolutional deep networks. In: Wang Q, Shi Y, Suk HI, Suzuki K (eds) Machine Learning in Medical Imaging. MLMI 2017, Lecture Notes in Computer Science, vol 10541, Springer, Cham. https://doi.org/10.1007/978-3-319-67389-9_44
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) MobileNetV2: inverted residuals and linear bottlenecks. The IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, USA, 18–23 June 2018, pp 4510–4520. https://doi.org/10.48550/arXiv.1801.04381
Seo H, Bassenne M, Xing L (2021) Closing the gap between deep neural network modeling and biomedical decision-making metrics in segmentation via adaptive loss functions. IEEE Trans Med Imaging 40:585–593. https://doi.org/10.1109/TMI.2020.3031913
Shillito LM (2018) Phytolith Analysis. In: López Varela SL (ed), The Encyclopedia of Archaeological Sciences. https://doi.org/10.1002/9781119188230.saseas0456
Soukup M, Martinka M, Cigáň M, Ravaszová F, Lux A (2014) New method for visualization of silica phytoliths in Sorghum bicolor roots by fluorescence microscopy revealed silicate concentration-dependent phytolith formation. Planta 240:1365–1372. https://doi.org/10.1007/s00425-014-2179-y
Strömberg CAE, Di Stilio VS, Song Z (2016) Functions of phytoliths in vascular plants: an evolutionary perspective. Funct Ecol 30:1286–1297. https://doi.org/10.1111/1365-2435.12692
Trembath-Reichert E, Wilson JP, McGlynn SE, Fischer WW (2015) Four hundred million years of silica biomineralization in land plants. Proc Natl Acad Sci 112:5449–5454. https://doi.org/10.1073/pnas.1500289112
Tsartsidou G, Lev-Yadun S, Efstratiou N, Weiner S (2009) Use of space in a Neolithic village in Greece (Makri): phytolith analysis and comparison of phytolith assemblages from an ethnographic setting in the same area. J Archaeol Sci 36:2342–2352. https://doi.org/10.1016/j.jas.2009.06.017
Tsutsui O, Sakamoto R, Obayashi M, Yamakawa S, Handa T, Nishio-Hamane D, Matsuda I (2016) Light and SEM observation of opal phytoliths in the mulberry leaf. Flora 218:44–50. https://doi.org/10.1016/j.flora.2015.11.006
Twiss PC, Suess E, Smith RM (1969) Morphological classification of grass phytoliths. Soil Sci Soc Am J 33:109–115. https://doi.org/10.2136/sssaj1969.03615995003300010030x
Valamoti SM (2009) Η αρχαιοβοτανική έρευνα της διατροφής στην προϊστορική Ελλάδα [The archaeobotanical research of nutrition in prehistoric Greece]. University Studio Press, Thessaloniki (in Greek)
Valamoti SM (2011) Ground cereal food preparations from Greece: the prehistory and modern survival of traditional Mediterranean ‘fast foods’. Archaeol Anthropol Sci 3:19–39. https://doi.org/10.1007/s12520-011-0058-z
Wang Z, Yang P, Liang H, Zheng C, Yin J, Tian Y, Cui W (2022) Semantic segmentation and analysis on sensitive parameters of forest fire smoke using Smoke-Unet and Landsat-8 Imagery. Remote Sens 14:45. https://doi.org/10.3390/rs14010045
Wendrich W, Ryan P (2012) Phytoliths and basketry materials at Çatalhöyük (Turkey): timelines of growth, harvest and objects life histories. Paléorient 38:55–63 (Prehistory of textiles in the Near East). https://www.jstor.org/stable/43264562
Wilson J, Hardy K, Allen R, Copeland L, Wrangham R, Collins M (2010) Automated classification of starch granules using supervised pattern recognition of morphological properties. J Archaeol Sci 37:594–604. https://doi.org/10.1016/j.jas.2009.10.024
Yao YF, Li X, Jiang HE, Ferguson DK, Hueber F, Ghosh R, Bera S, Li CS (2012) Pollen and phytoliths from fired ancient potsherds as potential indicators for deciphering past vegetation and climate in Turpan, Xinjiang, NW China. PLoS ONE 7:e39780. https://doi.org/10.1371/journal.pone.0039780
Zohary D, Hopf M, Weiss E (2012) Domestication of plants in the Old World, 4th edn. Oxford University Press, Oxford
Zurro D (2018) One, two, three phytoliths: assessing the minimum phytolith sum for archaeological studies. Archaeol Anthropol Sci 10:1673–1691. https://doi.org/10.1007/s12520-017-0479-4
Acknowledgements
The authors wish to thank Mr Evangelos Michelakis (agronomist) for generously offering his field for wheat cultivation at Chania, Crete, Greece; and Prof. Natàlia Alonso Martínez (University of Lleida) for generously offering the plant samples from Spain. The wheat cultivation in Spain was performed within the framework of the Experimental Camp of Protohistory (CEP) of Estinclells (Verdu, Urgell).
Prof. George E. Christidis (Technical University of Crete), Asst. Prof. Dan Cabanes (Rutgers, The State University of New Jersey), Dr Evangelia Kiriatzi (British School at Athens); and all the members of the Archaeobotany Unit (Catalan Institute of Human Paleoecology and Social Evolution) are gratefully acknowledged for making available the laboratory equipment and infrastructure necessary for the phytolith extraction and/or image acquisition procedure.
The editors and the reviewers of the Earth Science Informatics (ESIN) journal are acknowledged for their appreciated comments and remarks that have improved the manuscript. The Hellenic Academic Libraries Link (HEAL-Link) is sincerely appreciated for the financial support of the article publication in open access mode.
Funding
Open access funding provided by HEAL-Link (Greece).
Author information
Authors and Affiliations
Contributions
Conceptualisation: Panagiotis Partsinevelos, Nafsika C. Andriopoulou; Resources / Dataset: Nafsika C. Andriopoulou; Methodology: Georgios Petrakis, Nafsika C. Andriopoulou, Panagiotis Partsinevelos; Software: Georgios Petrakis; Validation: Georgios Petrakis; Original draft preparation: Nafsika C. Andriopoulou, Georgios Petrakis; Writing-Review and Editing: Nafsika C. Andriopoulou, Georgios Petrakis, Panagiotis Partsinevelos; Project administration / unding acquisition: Panagiotis Partsinevelos; Supervision: Panagiotis Partsinevelos.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Communicated by H. Babaie.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
ESM 1
(PDF 781 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Andriopoulou, N.C., Petrakis, G. & Partsinevelos, P. Twenty thousand leagues under plant biominerals: a deep learning implementation for automatic phytolith classification. Earth Sci Inform 16, 1551–1562 (2023). https://doi.org/10.1007/s12145-023-00975-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12145-023-00975-z