Abstract
Objectives
To assess the role of 16S ribosomal RNA analysis in microbial identification in febrile infants under six months of age diagnosed with UTI, and compare it with the conventional culture results.
Methods
Young infants under 6 mo of age who were suspected UTI from May 2018 to April 2019 had been enrolled. Uropathogens were analyzed by the traditional microbiologic culture system and the 16S rRNA analysis. The 16S rRNA analysis included 16S rRNA amplicon band confirmation and bacterial identification through the sequencing analysis.
Results
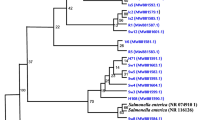
A total of 57 infants with the febrile UTI were enrolled, and the median age was 3 mo. Uropathogens were identified in 43 patients (75.4%) in a conventional culture method: Escherichia coli in 38 patients (88.4%), Klebsiella pneumoniae in 3 (7.0%), Enterobacter cloacae in 1 (2.3%), and Pseudomonas aeruginosa in 1 (2.3%). Fifty urine samples (87.8%) had positive 16S rRNA amplicon band on electrophoresis. Among the 16S rRNA–positive urines, 40 samples were available for the sequence analysis of 16S rRNA, and the identification of bacteria were as follows: E. fergusonii in 36, K. pneumoniae in 3, and Shigella flexneri in 1. The sensitivity of 16S rRNA sequencing was 81.4% [95% confidence interval (CI), 67.4–90.3%] and the specificity was 64.3% (95% CI, 38.8–83.7%).
Conclusion
Uropathogen identification using 16S rRNA analysis could be applied to manage the febrile UTI in young infants clinically in combination with the conventional culture.
Similar content being viewed by others
References
Srinivasan R, Karaoz U, Volegova M, et al. Use of 16S rRNA gene for identification of a broad rang of clinically relevant bacterial pathogens. PloS One. 2015;10:e0117617.
Chaudhari PP, Monuteaux MC, Bachur RG. Urine concentration and pyuria for identifying UTI in Infants. Pediatrics. 2016;138:e20162370.
Nickander KK, Shanholtzer CJ, Peterson LR. Urine culture transport tubes: effect of sample volume on bacterial toxicity of the preservative. J Clin Microbiol. 1982;15:593–5.
Tullus K. Difficulties in diagnosing urinary tract infections in small children. Pediatr Nephrol. 2011;26:1923–6.
Akram A, Maley M, Gosbell I, Nguyen T, Chavada R. Utility of 16S rRNA PCR performed on clinical specimens in patient management. Int J Infect Dis. 2017;57:144–9.
Yoo IY, Kang OK, Lee MK, et al. Comparison of 16S ribosomal RNA targeted sequencing and culture for bacterial identification in normally sterile body fluid samples: report of a 10-year clinical laboratory review. Ann Lab Med. 2020;40:63–7.
Patel JB. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol Diagn. 2001;6:313–21.
Yang B, Wang Y, Qian PY. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinformatics. 2016;17:135.
Davenport M, Mach KE, Shortliffe LMD, Banaei N, Wang TH, Liao JC. New and developing diagnostic technologies for urinary tract infections. Nat Rev Urol. 2017;14:296–310.
Elgaml A, Hassan R, Barwa R, Shokralla S, El-Naggar W. Analysis of 16S ribosomal RNA gene segments for the diagnosis of gram negative pathogenic bacteria isolated from urinary tract infections. Afr J Microbiol Res. 2013;7:2862–9.
Lehmann LE, Hauser S, Malinka T, et al. Rapid qualitative urinary tract infection pathogen identification by SeptiFast® real-time PCR. PLoS One.2011;6:e17146.
Straub J, Paula H, Mayr M, et al. Diagnostic accuracy of the ROCHE septifast PCR system for the rapid detection of blood pathogens in neonatal sepsis—a prospective clinical trial. PloS One. 2017;12:e0187688.
Roberts KB. Subcommittee on urinary tract infection, steering committee on quality improvement and management. Urinary tract infection: clinical practice guideline for the diagnosis and management of the initial UTI in febrile infants and children 2 to 24 months. Pediatrics. 2011;128:595–610.
Farmer JJ 3rd, Fanning GR, Davis BR, et al. Escherichia fergusonii and enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J Clin Microbiol. 1985;21:77–81.
Bours PH, Polak R, Hoepelman AI, Delgado E, Jarquin A, Matute AJ. Increasing resistance in community-acquired urinary tract infections in Latin America, five years after the implementation of national therapeutic guidelines. Int J Infect Dis. 2010;14:e770–4.
Lee JH. Discrimination of culture negative pyelonephritis in children with suspected febrile urinary tract infection and negative urine culture results. J Microbiol Immunol Infect. 2019;52:598–603.
Jaksic E, Bogdanovc R, Artiko V, et al. Diagnostic role of initial renal cortical scintigraphy in children with the first episode of acute pyelonephritis. Ann Nucl Med. 2011;25:37–43.
Hyun HS, Kim JH, Cho MH, et al. Low relapse rate of urinary tract infections from extended-spectrum beta-lactamase-producing bacteria in young children. Pediatr Nephrol. 2019;34:2399–407.
Petti CA, Polage CR, Schreckenberger P. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J Clin Microbiol. 2005;43:6123–5.
Barghouthi SA. A universal method for the identification of bacteria based on general PCR primers. Indian J Microbiol. 2011;51:430–44.
Funding
This study was supported by a grant from the Jeju National University Hospital Research Fund (2017).
Author information
Authors and Affiliations
Contributions
JHC was primarily responsible for conceptualization, methodology, data curation, and writing the original draft; YMY and YJK participated in software, validation, and composition of the work; KHH was involved in conceptualization, methodology, data curation, supervision, and writing, reviewing and editing the manuscript. KHH will act as the guarantor for this paper.
Corresponding author
Ethics declarations
Conflict of Interest
None.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Choi, J.H., Yoon, Y.M., Kim, YJ. et al. Role of 16S Ribosomal RNA Analysis in Identification of Microorganisms in Febrile Urinary Tract Infection of Infants. Indian J Pediatr 90, 660–664 (2023). https://doi.org/10.1007/s12098-022-04121-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12098-022-04121-3