Abstract
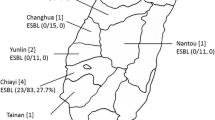
Extended-spectrum β-lactamase (ESBL) producing Escherichia coli represents a formidable challenge in the field of microbiology and public health due to its resistance to commonly used antibiotics. These strains pose a serious threat to human and animal health, underscoring the urgency of comprehensive research and surveillance. The ongoing investigation seeks ESBL producing E. coli strains from pig farms and slaughterhouses in West Bengal and Assam, India. A total of 309 samples were collected: nasal swabs (25), rectal swabs (25) from healthy pigs, pig pen soil (45), faeces (55), slaughterhouse effluents (115), and cleaning water (44). In these samples, 154 tested positive for E. coli, indicating a 49.8% prevalence. Among 154 E. coli isolates, 23 (14.9%) produced ESBLs, sourced from pig rectal swabs (7.1%), faeces (10.7%), slaughterhouse effluents (26.1%), and cleaning water (11.7%). Significantly, 4 ESBL E. coli isolates (6.6%) exclusively emerged from pig slaughterhouse effluents, displaying imipenem-resistant properties. The majority of ESBL E. coli primarily produced CTX-M and CMY, with consistent genetic markers blaCTX-M (100%) and blaCMY (82.6%). Remarkably, 2 (8.6%) of 17 ESBL E. coli isolates from pig slaughterhouse effluents carried the genetic marker blaNDM1. These findings stress implementing thorough surveillance in pig farms and local slaughterhouses. This proactive approach is crucial to identify ESBL E. coli strains, enhancing public health protection.
Similar content being viewed by others
References
Van Boeckel TP, Brower C, Gilbert M et al (2015) Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA 112:5649–5654. https://doi.org/10.1073/pnas.1503141112
Mutua F, Sharma G, Grace D et al (2020) A review of animal health and drug use practices in India, and their possible link to antimicrobial resistance. Antimicrob Resist Infect Control 9:1–13
Czekalski N, Berthold T, Caucci S et al (2012) Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front Microbiol. https://doi.org/10.3389/fmicb.2012.00106
Dcosta VM, King CE, Kalan L et al (2011) Antibiotic resistance is ancient. Nature 477:457–461
Baquero F, Martínez JL, Cantón R (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265
Søraas A, Sundsfjord A, Sandven I et al (2013) Risk factors for community-acquired urinary tract infections caused by ESBL-producing enterobacteriaceae—a case-control study in a low prevalence country. PLoS ONE. https://doi.org/10.1371/journal.pone.0069581
Njage PMK, Buys EM (2015) Pathogenic and commensal Escherichia coli from irrigation water show potential in transmission of extended spectrum and AmpC β-lactamases determinants to isolates from lettuce. Microb Biotechnol 8:462–473. https://doi.org/10.1111/1751-7915.12234
Solà-Ginés M, González-López JJ, Cameron-Veas K et al (2015) Houseflies (Musca domestica) as vectors for extended-spectrum β-lactamase-producing Escherichia coli on Spanish broiler farms. Appl Environ Microbiol 81:3604–3611. https://doi.org/10.1128/AEM.04252-14
North J (2020) Challenges to tackling antimicrobial resistance. Cambridge University Press, Cambridge
Gao LL, Tan Y, Zhang XD et al (2015) Emissions of Escherichia coli carrying extended-spectrum β-lactamase resistance from pig farms to the surrounding environment. Int J Environ Res Public Health 12:4203–4213. https://doi.org/10.3390/ijerph120404203
Von Salviati C, Laube H, Guerra B et al (2015) Emission of ESBL/AmpC-producing Escherichia coli from pig fattening farms to surrounding areas. Vet Microbiol 175:77–84. https://doi.org/10.1016/j.vetmic.2014.10.010
Samanta A, Mahanti A, Chatterjee S et al (2018) Pig farm environment as a source of beta-lactamase or AmpC-producing Klebsiella pneumoniae and Escherichia coli. Ann Microbiol 68:781–791. https://doi.org/10.1007/s13213-018-1387-2
Chen X, Han W, Patel M et al (2022) Inactivation of a pathogenic NDM-1-positive Escherichia coli strain and the resistance gene blaNDM-1 by TiO2/UVA photocatalysis. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2022.157369
Deb R, Chaudhary P, De S (2022) CRISPR/cas9 cassette targeting Escherichia coli blaCTX-M specific gene of mastitis cow milk origin can alter the antibiotic resistant phenotype for cefotaxime. Anim Biotechnol. https://doi.org/10.1080/10495398.2022.2053695
Wang GCY, Wang Y (1996) The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142:1107–1114. https://doi.org/10.1099/13500872-142-5-1107
Gālina D, Balins A, Valdovska A (2021) The prevalence and characterization of fecal extended-spectrum-beta-lactamase-producing Escherichia coli isolated from pigs on farms of different sizes in Latvia. Antibiotics 10:1099. https://doi.org/10.3390/antibiotics10091099
Drieux L, Brossier F, Sougakoff W, Jarlier V (2008) Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect 14:90–103
Lalzampuia H, Dutta TK, Warjri I, Chandra R (2013) PCR-based detection of extended-spectrum β-lactamases (blaCTX-M-1and blaTEM) in Escherichia coli, Salmonella spp. and Klebsiella pneumoniae Isolated from Pigs in North Eastern India (Mizoram). Indian J Microbiol 53:291–296. https://doi.org/10.1007/s12088-013-0378-z
Machado E, Coque TM, Cantón R et al (2008) Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J Antimicrob Chemother 62:296–302. https://doi.org/10.1093/jac/dkn179
Duan RS, Sit THC, Wong SSY et al (2006) Escherichia coli producing CTX-M β-lactamases in food animals in Hong Kong. Microb Drug Resist 12:145–148. https://doi.org/10.1089/mdr.2006.12.145
Geser N, Stephan R, Hächler H (2012) Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet Res. https://doi.org/10.1186/1746-6148-8-21
Hiroi M, Yamazaki F, Harada T et al (2012) Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J Vet Med Sci 74:189–195. https://doi.org/10.1292/jvms.11-0372
Mirbagheri SZ, Meshkat Z, Naderinasab M et al (2015) Study on imipenem resistance and prevalence of blaVIM1 and blaVIM2 metallo-beta lactamases among clinical isolates of Pseudomonas aeruginosa from Mashhad, Northeast of Iran. Iran J Microbiol 7:72–78
Nandi SP, Sultana M, Hossain MA (2013) Prevalence and characterization of multidrug-resistant zoonotic Enterobacter spp. in Poultry of Bangladesh. Foodborne Pathog Dis 10:420–427. https://doi.org/10.1089/fpd.2012.1388
Yaici L, Haenni M, Saras E et al (2016) blaNDM-5-carrying IncX3 plasmid in Escherichia coli ST1284 isolated from raw milk collected in a dairy farm in Algeria. J Antimicrob Chemother 71:2671–2672
Fischer J, San José M, Roschanski N et al (2017) Spread and persistence of VIM-1 Carbapenemase-producing Enterobacteriaceae in three German swine farms in 2011 and 2012. Vet Microbiol 200:118–123. https://doi.org/10.1016/j.vetmic.2016.04.026
Pruthvishree BS, Vinodh Kumar OR, Sinha DK et al (2017) Spatial molecular epidemiology of carbapenem-resistant and New Delhi metallo beta-lactamase (blaNDM)-producing Escherichia coli in the piglets of organized farms in India. J Appl Microbiol 122:1537–1546. https://doi.org/10.1111/jam.13455
Pulss S, Semmler T, Prenger-Berninghoff E et al (2017) First report of an Escherichia coli strain from swine carrying an OXA-181 carbapenemase and the colistin resistance determinant MCR-1. Int J Antimicrob Agents 50:232–236. https://doi.org/10.1016/j.ijantimicag.2017.03.014
Shi X, Li Y, Yang Y et al (2021) High prevalence and persistence of carbapenem and colistin resistance in livestock farm environments in China. J Hazard Mater. https://doi.org/10.1016/j.jhazmat.2020.124298
Dankittipong N, Fischer EAJ, Swanenburg M et al (2022) Quantitative risk assessment for the introduction of carbapenem-resistant Enterobacteriaceae (CPE) into Dutch Livestock Farms. Antibiotics. https://doi.org/10.3390/antibiotics11020281
Jia B, Raphenya AR, Alcock B et al (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. https://doi.org/10.1093/nar/gkw1004
Ewers C, Bethe A, Semmler T et al (2012) Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect 18:646–655
Hu YY, Cai JC, Zhou HW et al (2013) Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl Environ Microbiol 79:5988–5996. https://doi.org/10.1128/AEM.01740-13
Rodríguez I, Barownick W, Helmuth R et al (2009) Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003–07. J Antimicrob Chemother 64:301–309. https://doi.org/10.1093/jac/dkp195
Mollenkopf DF, Kleinhenz KE, Funk JA et al (2011) Salmonella enterica and Escherichia coli harboring blaCMY in retail beef and pork products. Foodborne Pathog Dis 8:333–336. https://doi.org/10.1089/fpd.2010.0701
Heider LC, Hoet AE, Wittum TE et al (2009) Genetic and phenotypic characterization of the blaCMY gene from Escherichia coli and Salmonella enterica isolated from food-producing animals, humans, the environment, and retail meat. Foodborne Pathog Dis 6:1235–1240. https://doi.org/10.1089/fpd.2009.0294
Evans BA, Amyes SGB (2014) OXA β-lactamases. Clin Microbiol Rev 27:241–263. https://doi.org/10.1128/CMR.00117-13
Sugumar M, Kumar KM, Manoharan A et al (2014) Detection of OXA-1 β-lactamase gene of Klebsiella pneumoniae from blood stream infections (BSI) by conventional PCR and in-silico analysis to understand the mechanism of OXA mediated resistance. PLoS One 9:e91800. https://doi.org/10.1371/journal.pone.0091800
Zhou K, Lokate M, Deurenberg RH et al (2015) Characterization of a CTX-M-15 producing Klebsiella pneumoniae outbreak strain assigned to a novel sequence type (1427). Front Microbiol. https://doi.org/10.3389/fmicb.2015.01250
Mir RA, Weppelmann TA, Johnson JA et al (2016) Identification and characterization of cefotaxime resistant bacteria in beef cattle. PLoS ONE. https://doi.org/10.1371/journal.pone.0163279
Walsh TR, Weeks J, Livermore DM, Toleman MA (2011) Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. https://doi.org/10.1016/S1473-3099(11)70059-7
Zhai R, Fu B, Shi X et al (2020) Contaminated in-house environment contributes to the persistence and transmission of NDM-producing bacteria in a Chinese poultry farm. Environ Int. https://doi.org/10.1016/j.envint.2020.105715
Acknowledgements
Authors acknowledge the financial support received from Indian Council of Agricultural Research, New Delhi, India under the project of Indian Network for Fisheries and Animal Antimicrobial Resistance (INFAAR). Authors are thankful to Director, ICAR-National Research Centre on Pig, Guwahati, Assam, India for providing necessary facilities to conduct the experiment.
Funding
Authors acknowledge the financial support received from Indian Council of Agricultural Research, New Delhi, India under the project of Indian Network for Fisheries and Animal Antimicrobial Resistance (INFAAR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declared that there is no conflict of interest for the present study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Niharika, J., Deb, R., Parihar, R. et al. Isolation and Characterization of Extended-Spectrum β-Lactamase Producing Escherichia coli from Pig Farms and Slaughterhouse. Indian J Microbiol (2023). https://doi.org/10.1007/s12088-023-01151-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12088-023-01151-z