Abstract
Increasing evidences have illustrated the important role of N6-methyladenosine (m6A) in atherosclerosis (AS). However, the role of m6A modification in AS pathophysiological process is still unknown. Here, the present work tried to investigate the expression and function of m6A methyltransferase KIAA1429 in AS pathology and explored its undergoing m6A-dependent molecular mechanism. Results indicated that KIAA1429 remarkedly up-regulated in oxidative low-density lipoprotein (ox-LDL)-treated human umbilical vein endothelial cells (HUVECs). KIAA1429 over-expression inhibited the proliferation/migration in ox-LDL-treated HUVECs, while, KIAA1429 knockdown up-regulated the proliferation and migration. Mechanistically, via m6A modification sites binding, ROCK2 mRNA was post-transcriptionally upregulated by KIAA1429 in response to Actinomycin D. Collectively, our study demonstrated the regulation of KIAA1429 on ox-LDL-induced HUVECs via m6A/ROCK2 pathway. These findings provide new insights for m6A-mediated epigenetics in AS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Atherosclerosis (AS) acts as chronic inflammatory vascular disorders with increasing morbidity worldwide, which is a deposition of plaque accumulation and multifactorial inflammatory process in vascular walls [1, 2]. AS is responsible for occurrence of various clinical manifestation, e.g., coronary heart diseases, peripheral arterial disease and myocardial infarction and stroke [3, 4]. Low density lipoprotein (LDL) functions as a basic factor of AS. LDL enters into the vascular wall through vascular endothelium, and the retained LDL is modified into ox-LDL. Ox-LDL is phagocytosed by macrophages to form foam cells, which continuously increases and fuse to form the lipid core of AS plaque [5, 6]. Endothelial dysfunctions, especially endothelial cells damage, function as critical pathology for AS [7, 8].
N6-methyladenosine (m6A) acts as the most abundant posttranscriptional modification for mRNAs, which partially determines the fate of RNA, including RNA stabilization, splicing, and nuclear export [9, 10]. m6A methylations have been shown to regulate series of inflammatory processes, including the inflammatory vascular disorder of endothelial cells in AS [11]. For example, FTO-mediated m6A demethylations regulate the expression of lipid-related genes and regulates lipid metabolism to lead to occurrence of diabetic hyperlipidemia [12]. m6A methyltransferase methyltransferase-like 3 (METTL3) is highly expressed in ox-LDL-induced HUVECs and METTL3 knockdown inhibits HUVECs’ proliferation and tube formation in ox-LDL-treated HUVECs. Besides, METTL3 positively regulates JAK2/STAT3 pathway in m6A-dependent manner in HUVECs [13]. Therefore, the emerging findings indicate the critical roles of m6A in AS.
Given that the potential function of m6A in abnormal lipids metabolism-related AS pathology, we focused on the undergoing regulation of m6A in ox-LDL-induced HUVECs. In the initial screening, we detected several m6A key enzymes (i.e., KIAA1429, METTL3, WTAP) to discovery the up-regulated or down-regulated elements in ox-LDL-induced HUVECs. Results showed that the novel m6A writer KIAA1429 up-regulated upon ox-LDL administered. Finally, we focused on KIAA1429 and then explored its functions. Our cellular assays’ results showed that the expression of KIAA1429 (also known as VIRMA) increased upon ox-LDL administered and the response showed a dosage-dependent manner. KIAA1429 could regulate the proliferation and migration of HUVECs in ox-LDL administered. Moreover, KIAA1429 bound to the m6A modification sites of ROCK2 mRNA and then enhanced its mRNA stability. In conclusion, m6A writer KIAA1429 targeted m6A/ROCK2 axis to regulate the proliferation/migration of endothelial cells.
Materials and Methods
Cell Culture
HUVECs (Human umbilical vein endothelial cells) cell lines were provided by the Institute of the Chinese Academy of Sciences (Shanghai, China). Then, the cells were cultured in endothelial cell medium (Catalog #1001, ScienCell Research Laboratories, San Diego, CA, United States) supplemented with 10–12% fetal bovine serum (FBS, Catalog #0500, ScienCell Research Laboratories), 1% endothelial cell growth supplement (ECGS, Catalog #1052, ScienCell Research Laboratories), and 1% penicillin/streptomycin solution (Catalog #0513, ScienCell Research Laboratories). Passages 2–4 of cells were used for this project. All cells were cultured in humidified air in 37 °C at 5% CO2. ox-LDL (Yesen, Shanghai, China) was added to HUVECs (0–100 μg/mL concentration, 24 h) to construct endothelial cell injury. This study was approved by the ethics committee of Tianjin Hospital.
Transfection
For the silencing of KIAA1429 in vitro studies, the shRNA targeting KIAA1429 (sh-KIAA1429) and negative control siRNA (sh-NC) were synthesized (RiboBio, Guangzhou, China) based on the manufacturer’s suggestion. As regarding to overexpression of KIAA1429, pcDNA based overexpression plasmid specific to KIAA1429 or corresponding scrambled oligonucleotide sequences as a negative control were transfected.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from HUVECs samples using modified TRIzol co-purification technique (Invitrogen, Hemel Hempstead, UK) as previously reported [14]. In brief, 500 μl of cell suspension was washed by 75–78% ethanol before solubilization using nuclease-free water (50 μl). To identify the level of KIAA1429 mRNA, RNA (1 μg) was reversely transcribed to cDNA by PrimeScript™ RT Reagent Kit (TaKaRa). Then, RT-qPCR was performed using One Step TB Green® PrimeScript™ PCR Kit (TaKaRa, Cat.#RR086B) as recommended by manufacturers’ protocols. The quantification of mRNA reference gene was calculated using the 2−ΔΔCt method. The primers used in PCR amplification were showed in Table S1.
Western Blot
Total proteins were collected using RIPA buffer (Thermo Scientific, CA, USA) and then quantified for protein quantification by using Pierce™ BCA Protein Quantification Kit (Thermo Scientific) [15]. Protein (20 μg) was used for electrophoresis loading SDS-PAGE and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% skimmed milk and incubated with primary antibodies, including anti-KIAA1429 (Cell Signaling Technology, 1:1000, #88,358), anti-ROCK2 (Cell Signaling Technology, 1:1000, #47,012), and beta-actin (CWBio, Beijing, China). After incubation by primary antibodies or their corresponding secondary antibodies, blots were developed using SuperSignal West Dura Persistence Substrate (Thermo Scientific).
Proliferation CCK-8 Assay
For the proliferation of HUVECs, CCK-8 assay was performed. HUVECs cells were plated onto 96-well plates. 10μL CCK-8 reagent (Dojindo Japan) was added into each well at 24, 48, 72, and 96 h. Then, optical density (OD) value of wells was detected at 450 nm was recorded by automatic enzyme-mark reader (Multiskan FC, Thermo Fisher Scientific, Waltham, MA, USA) using a microplate reader.
Migration Assay
For the migration of HUVECs, wound healing assay was performed. In brief, 2 × 104 HUVECs about 90% confluence were plated 6-well plates. Medium was removed and then cell monolayers were manually wounded with 200 ul pipette tip. After twice washing with PBS, cells were incubated at 37 °C. The wound closure was evaluated with an inverted microscope. The migration rate was quantified according to distance. The migration rate was calculated: migration rate = migration distance/original distance.
m6A Quantification
The m6A quantification of global mRNA was measured by EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) (Epigentek) following the manufacturer’s protocol. 200 ng Poly-A-purified RNA was coated on assay wells and capture antibody solution was separately added to suitable concentration following the manufacturer’s instructions. The m6A level was colorimetrically quantified by absorbance at a wavelength of 450 nm.
RNA Immunoprecipitation (RIP) assay
The interaction within RNAs was identified by RIP-PCR. RIP assay was performed by Magna RIP-RNA-Binding Protein Immunoprecipitation Kit (Magna RIP Kit; Millipore, Bedford, MA, USA). Cell was lysed in lysis buffer with protease inhibitor/RNase inhibitor, and the cell extract was incubated with protein A/G agarose beads conjugated with antibody/IgG at 4 °C for 2 h. Magnetic dynabeads (Life Technologies, USA) were washed or incubated with Proteinase K. Finally, purified RNA was subjected for RT-PCR analysis.
m.6A-RNA Immunoprecipitation PCR (MeRIP-PCR)
To quantify the m6A-modified ROCK2 mRNA, MeRIP-PCR was performed. In brief, anti-m6A antibody (Millipore, cat. #ABE572) was conjugated to protein A/G magnetic beads in IP buffer (20 mM Tris pH 7.5, 1% NP-40, 140 mM NaCl, 2 mM EDTA). Total RNA was isolated from HUVECs and incubated with the antibody in IP buffer supplemented with RNase inhibitor and protease inhibitor. The precipitated RNA was eluted with elution buffer and detected using further qRT-PCR assay.
RNA Stability
To analyze the ROCK2 mRNA stability, HUVECs were administrated with actinomycin D (Act D, 2 μg/mL, Sigma) and then collected in different time-point (0 h, 3 h, 6 h). ROCK2 mRNAs were extracted using Trizol reagent. Finally, mRNA levels were detected by qRT-PCR and normalized to the values measured in the mock treatment group (the 0 h group) using the primers: forward, 5’- TCAGAGGTCTACAGATGAAGGC-3′, reverse, 5′- CCAGGGGCTATTGGCAAAGG-3′.
Statistical Analysis
Data was expressed as the mean ± standard deviation (SD) in independent replicate. Student’s t-test or variance (ANOVA) analysis was used to compare difference of independent groups. Assays were performed in triplicate. Data were exhibited as Mean ± Standard Deviation (SD). **p < 0.01, *p < 0.05 were considered a statistical difference.
Results
KIAA1429 up-Regulated in the ox-LDL-Induced HUVECs
To construct the cellular AS model, human endothelial cells (HUVECs) were administrated with ascending ox-LDL (0–100 μg/mL) concentration. With the ascending dosage of ox-LDL, m6A quantitative analysis was performed and results demonstrated that m6A modification level increased upon ox-LDL administered (Fig. 1A). Besides, the level of m6A writer KIAA1429 in the ox-LDL-induced HUVECs was detected and results indicated that KIAA1429 mRNA (Fig. 1B) and protein levels (Fig. 1C) were upregulated in ox-LDL treated HUVECs. Moreover, the proliferation of HUVECs was detected and results illustrated that proliferative ability of HUVECs was repressed in ox-LDL administered in dosage-dependent manner (Fig. 1D). In conclusion, these results and findings suggested that KIAA1429 upregulated in ox-LDL-induced HUVECs.
KIAA1429 up-regulated in the ox-LDL-induced HUVECs. A m6A quantitative analysis was performed to detect the m6A modification level in HUVECs with ox-LDL administered (0–100 μg/mL). B RT-PCR detected the level of KIAA1429 mRNA in HUVECs with ox-LDL administered (0–100 μg/mL). C Western blot was performed to show the KIAA1429 protein in HUVECs with ox-LDL administered (0–100 μg/mL). D The proliferation of HUVECs was detected using CCK-8 to illustrate the proliferative ability of HUVECs. Assays were performed in triplicate. Data were exhibited as Mean ± Standard Deviation (SD). **p < 0.01, *p < 0.05
KIAA1429 Inhibited the Proliferation/Migration for ox-LDL Administrated HUVECs
Given that KIAA1429 showed an overexpression in ox-LDL administrated human endothelial cells (HUVECs), we subsequently performed the bio-functional assays to confirm the role of KIAA1429 in AS. Firstly, KIAA1429 knockdown (Fig. 2A) and overexpression (Fig. 2B) transfection were respectively constructed in HUVECs. The proliferation of HUVECs was detected and results illustrated that proliferative ability of HUVECs was up-regulated in KIAA1429 knockdown, while KIAA1429 overexpression repressed proliferation (Fig. 2C). Wound healing assay was performed and results indicated that KIAA1429 knockdown promoted the migration of HUVECs, while KIAA1429 overexpression reduced it (Fig. 2D). In conclusion, these results and findings suggested that KIAA1429 inhibited the proliferation/migration for ox-LDL administrated HUVECs.
KIAA1429 repressed the proliferation/migration for ox-LDL administrated HUVECs. A, B RT-qPCR and western blot for (A) KIAA1429 knockdown and (B) KIAA1429 overexpression were performed to detect the efficient of transfection. C CCK-8 assay for the proliferation of HUVECs was performed to illustrate the proliferative ability of HUVECs upon KIAA1429 knockdown and KIAA1429 overexpression. D Wound healing assay indicated the migration of HUVECs upon KIAA1429 knockdown and KIAA1429 overexpression. Assays were performed in triplicate. Data were exhibited as Mean ± Standard Deviation (SD). **p < 0.01, *p < 0.05
ROCK2 Acted as the Target of KIAA1429
Using diverse screening methods, we carefully discovered the target of KIAA1429, e.g., bioinformatics predictive analysis and multiplex cell assays (RT-PCR, western blot et. al). Our pre-experiments data illustrated that ROCK2 (Rho associated coiled-coil containing protein kinase 2) had m6A modification sites in its 3’-UTR, which suggested that ROCK2 might act as the target of KIAA1429 (Fig. 3A). In the pathophysiology of AS, ROCK2 functions as an essential element in AS [16]. The m6A modification site on ROCK2 mRNA was ‘GGACU’ motif. In the ROCK2 gene, the m6A modification site distributed in 3’-UTR of ROCK2 (Fig. 3B). Using the SRAMP online tool (http://www.cuilab.cn/sramp), analysis demonstrated that there were m6A modification site on ROCK2 mRNA (Fig. 3C). In ox-LDL-treated HUVECs, m6A quantitative analysis indicated that KIAA1429 knockdown reduced the m6A modification level, while KIAA1429 overexpression up-regulated the m6A modification level (Fig. 3D). RIP-PCR analysis revealed that KIAA1429 remarkably interacted with ROCK2 mRNA in HUVECs (Fig. 3E). In conclusion, these results and findings suggested that ROCK2 acted as the target of KIAA1429.
ROCK2 acted as the target of KIAA1429. A The m6A modification site on ROCK2 (Rho associated coiled-coil containing protein kinase 2) mRNA was ‘GGACU’ motif. B In the ROCK2 gene, the m6A modification site distributed in 3’-UTR of ROCK2. C SRAMP online tool (http://www.cuilab.cn/sramp) demonstrated the m6A modification site on ROCK2 mRNA. D m6A quantitative analysis was performed to detect the role of KIAA1429 knockdown/overexpression on the ox-LDL-treated HUVECs. E RIP-qPCR analysis indicated interaction within KIAA1429 and ROCK2 mRNA in HUVECs. Assays were performed in triplicate. Data were exhibited as Mean ± Standard Deviation (SD). **p < 0.01, *p < 0.05
KIAA1429 Enhanced the Stability of ROCK2 mRNA
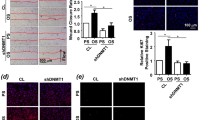
Moreover, we tried to explore the function of KIAA1429 on ROCK2. Firstly, MeRIP-PCR analysis was performed to detect the m6A modification level on ROCK2 mRNA. Results indicated that KIAA1429 knockdown reduced the m6A modification on ROCK2 mRNA (Fig. 4A), while KIAA1429 overexpression up-regulated the m6A modification level (Fig. 4B). In the ox-LDL-induced HUVECs, ROCK2 mRNA increased upon ox-LDL administered in dosage-dependent manner (Fig. 4C). RIP-qPCR analysis found that KIAA1429 knockdown reduced the precipitated ROCK2 mRNA enrichment, while KIAA1429 overexpression up-regulated the precipitated ROCK2 mRNA enrichment (Fig. 4D). RNA stability analysis revealed that KIAA1429 knockdown decreased the ROCK2 mRNA upon Act D treatment, while KIAA1429 overexpression up-regulated the ROCK2 mRNA upon Act D treatment (Fig. 4E). Moreover, KIAA1429 knockdown reduced the ROCK2 protein in ox-LDL-induced HUVECs, while KIAA1429 overexpression increased the ROCK2 protein (Fig. 4F). In conclusion, these results and findings suggested that KIAA1429 enhanced the stability of ROCK2 mRNA.
KIAA1429 enhanced the stability of ROCK2 mRNA. A MeRIP-PCR analysis was performed using the anti-m6A antibody to detect the m6A modification level on ROCK2 mRNA. Ox-LDL-induced HUVECs were transfected with KIAA1429 knockdown (sh-NC-IgG, sh-NC-m6A, sh-KIAA1429-IgG, sh-KIAA1429- m6A). B Ox-LDL-induced HUVECs were transfected with KIAA1429 overexpression (vector-IgG, vector-m6A, KIAA1429-IgG, KIAA1429- m6A). C RT-PCR was performed to detect the ROCK2 mRNA level in the ox-LDL-induced HUVECs. D RIP-qPCR analysis was performed using anti-m6A antibody. The precipitated ROCK2 mRNA enrichment was analyzed as compared to controls. E RNA stability analysis was performed to detect the ROCK2 mRNA upon Act D treatment in ox-LDL-induced HUVECs. F Western blot analysis was performed to detect the ROCK2 protein in ox-LDL-induced HUVECs. Assays were performed in triplicate. Data were exhibited as Mean ± Standard Deviation (SD). **p < 0.01, *p < 0.05
Discussion
In atherosclerosis (AS), the activity of vascular endothelial cells could reflect cells’ metabolism and proliferation [17,18,19]. AS is a life-threatening vascular disease, and m6A modification level is dysregulated in its pathophysiologic processes of AS [20, 21]. Here, we found that m6A writer KIAA1429 upregulated in ox-LDL-induced HUVECs. Moreover, the function and corresponding molecular mechanism in AS progression are of great value for precision targeted therapy.
Here, this research constructed cellular AS model using ox-LDL-induced HUVECs, and results revealed that m6A level increased upon ox-LDL administered. Because the deregulated m6A level in ox-LDL-induced HUVECs, we assumed that m6A regulators might participate in HUVECs’ pathophysiology.
In the initial screening, we detected several m6A key enzymes (i.e., KIAA1429, METTL3, WTAP) to discovery the up-regulated or down-regulated elements. Finally, we focused on the novel m6A writer KIAA1429 and then explored its functions. Our cellular assays’ results showed that the expression of KIAA1429 increased upon ox-LDL administered and the response showed a dosage-dependent manner.
Functional assays illustrated that KIAA1429 overexpression repressed proliferation and migration of HUVECs, while KIAA1429 knockdown recovered the repression on HUVECs’ proliferation and migration. As is known to all that the ability of proliferation/migration of vascular endothelial cell is very critical for the vascular bio-function. Thus, in these findings, KIAA1429 regulated the proliferation and migration of HUVECs, which significantly showed the roles of KIAA1429 on HUVECs.
Increasing evidence synergistically indicates the potential vital roles of m6A modification on AS [22]. For instance, ox-LDL remarkably stimulation promotes the m6A modification level of macrophages in AS and METTL3 knockdown inhibits the oxLDL-induced inflammatory response and m6A modification [23]. Besides, ALKBH5 low-expression significantly increases SPHK1 m6A mRNA methylation, in contrast, METTL3 overexpression reduces expression of SPHK1 mRNA [24]. In vascular endothelium of atherogenic inflammatory cascades, METTL3-mediated RNA hypermethylation up-regulates NLRP1 mRNA transcript and down-regulates KLF4 transcript via YTHDF1/YTHDF2 m6A reader proteins [25]. Thus, these findings suggest the vital functions of m6A on AS.
The online tool suggested that there were m6A modification sites on ROCK2 mRNA (Fig. 3C). Moreover, we performed RIP-PCR (Fig. 3E, 4D) and MeRIP-PCR (Fig. 4A, B) to identify the molecular interaction within KIAA1429 and ROCK2 mRNA. In the pathophysiology of AS, ROCK2 functions as an essential element in AS [16]. For instance, circCHMP5/ROCK2 axis regulates cell cycle, proliferation, angiogenesis and inflammation in ox-LDL-induced HUVECs [26]. CircUSP36/ROCK2 axis regulates cell apoptosis and inflammatory responses, and promotes cell migration and invasion in ox‑LDL‑induced injury for HUVECs [27]. Here, we found that ROCK2 up-regulated in the ox-LDL-induced HUVECs. KIAA1429 targeted the m6A modification site of ROCK2 mRNA and then up-regulated the mRNA stability of ROCK2. Therefore, in this results, KIAA1429/ROCK2 axis accelerated the proliferation and migration of HUVECs.
For the deficiencies and defects, we talk briefly about what we’ve found in this research. Firstly, being limited by the laboratory external conditions, in vivo assays were unable to proceed as we initially assumed. Moreover, clinical sample research was difficult to put into practice due to the COVID-19. Furthermore, our understanding of m6A is still in its infancy because of the absence of more research. With the great development of m6A and Epigenomics, it would be greatly help to the contribution of our work to the field of AS.
Conclusion
In conclusion, the present research found a novel manner in AS by which KIAA1429 negatively regulated the proliferation and migration of HUVECs. Mechanistically, KIAA1429 targeted the m6A modification site of ROCK2 mRNA to install m6A modification of ROCK2, thereby enhancing ROCK2 mRNA stability (Fig. 5). These new findings provide novel insight for vascular endothelial cells injury for AS.
Data Availability
No research data shared.
References
Keeter, W. C., Ma, S., Stahr, N., Moriarty, A. K., & Galkina, E. V. (2022). Atherosclerosis and multi-organ-associated pathologies. Seminars in Immunopathology, 44, 363–374.
Kong, P., Cui, Z. Y., Huang, X. F., Zhang, D. D., Guo, R. J., & Han, M. (2022). Inflammation and atherosclerosis: Signaling pathways and therapeutic intervention. Signal Transduction and Targeted Therapy, 7, 131.
Liu, D., Liu, J., Zhang, D., & Yang, W. (2022). Advances in relationship between cell senescence and atherosclerosis. Zhejiang da xue xue bao Yi xue ban = Journal of Zhejiang University Medical sciences., 51, 95–101.
Tuvali, O., Sella, G., Haberman, D., Cuciuc, V., & George, J. (2022). Colchicine in Atherosclerotic Vascular Disease: The Good, the Bad, and the Ugly. The Israel Medical Association Journal : IMAJ, 24, 191–197.
Chan, Y. H., & Ramji, D. P. (2022). Atherosclerosis: pathogenesis and key cellular processes current and emerging therapies, key challenges, and future research directions. Methods in Molecular Biology, 2419, 3–19.
Kaiser, Y., Daghem, M., Tzolos, E., Meah, M. N., Doris, M. K., Moss, A. J., Kwiecinski, J., Kroon, J., Nurmohamed, N. S., van der Harst, P., Adamson, P. D., Williams, M. C., Dey, D., Newby, D. E., Stroes, E. S. G., Zheng, K. H., & Dweck, M. R. (2022). Association of Lipoprotein(a) With Atherosclerotic Plaque Progression. Journal of the American College of Cardiology, 79, 223–233.
Kłosiewicz-Latoszek, L., Cybulska, B., Stoś, K., & Tyszko, P. (2021). Hypolipaemic nutraceutics: Red yeast rice and Armolipid, berberine and bergamot. Annals of Agricultural and Environmental Medicine : AAEM, 28, 81–88.
Madeline Chee, Y. M., Lew, P. S., & Darryl Lim, M. J. (2020). True Idiopathic Radial Artery Aneurysm: A Case Report and Review of Current Literature. EJVES Vascular Forum, 49, 34–39.
Chang, Y., Yi, M., Wang, J., Cao, Z., Zhou, T., Ge, W., Muhammad, Z., Zhang, Z., Feng, Y., Yan, Z., Felici, M., Shen, W., & Cao, H. (2022). Genetic regulation of N6-methyladenosine-RNA in mammalian gametogenesis and embryonic development. Frontiers in Cell and Developmental Biology, 10, 819044.
Li, Y., Meng, L., & Zhao, B. (2022). The roles of N6-methyladenosine methylation in the regulation of bone development, bone remodeling and osteoporosis. Pharmacology & Therapeutics, 238, 108174.
Wu, W., Zhang, F., Zhao, J., He, P., & Li, Y. (2022). The N6-methyladenosine:Mechanisms, diagnostic value, immunotherapy prospec-ts and challenges in gastric cancer. Experimental cell Research, 415, 113115.
Yang, Z., Yu, G. L., Zhu, X., Peng, T. H., & Lv, Y. C. (2022). Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes & diseases, 9, 51–61.
Dong, G., Yu, J., Shan, G., Su, L., Yu, N., & Yang, S. (2021). N6-methyladenosine methyltransferase METTL3 promotes angiogenesis and atherosclerosis by upregulating the JAK2/STAT3 pathway via m6A reader IGF2BP1. Frontiers in Cell and Developmental Biology, 9, 731810.
Jia, J., Wang, Y., Huang, R., Du, F., Shen, X., Yang, Q., & Li, J. (2022). Protein disulfide-isomerase A3 knockdown attenuates oxidized low-density lipoprotein-induced oxidative stress, inflammation and endothelial dysfunction in human umbilical vein endothelial cells by downregulating activating transcription factor 2. Bioengineered, 13, 1436–1446.
Xu, L., Xu, C., Lin, X., Lu, H., & Cai, Y. (2021). Interference with lysophosphatidic acid receptor 5 ameliorates oxidized low-density lipoprotein-induced human umbilical vein endothelial cell injury by inactivating NOD-like receptor family, pyrin domain containing 3 inflammasome signaling. Bioengineered, 12, 8089–8099.
Takeda, Y., Matoba, K., Kawanami, D., Nagai, Y., Akamine, T., Ishizawa, S., Kanazawa, Y., Yokota, T., & Utsunomiya, K. (2019). ROCK2 Regulates Monocyte Migration and Cell to Cell Adhesion in Vascular Endothelial Cells. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms20061331
Zheng, L., Li, M., Wei, J., Chen, S., Xue, C., Zhan, Y., Duan, Y., Deng, H., Xiong, W., Li, G., & Zhou, M. (2022). The emerging roles of the interaction between m6A modification and c-Myc in driving tumorigenesis and development. Journal of Cellular Physiology. https://doi.org/10.1002/jcp.30733
Chang, H., Yang, J., Wang, Q., Zhao, J., & Zhu, R. (2022). Role of N6-methyladenosine modification in pathogenesis of ischemic stroke. Expert Review of Molecular Diagnostics. https://doi.org/10.1080/14737159.2022.2049246
Chen, Y. S., Ouyang, X. P., Yu, X. H., Novák, P., Zhou, L., He, P. P., & Yin, K. (2021). N6-Adenosine Methylation (m(6)A) RNA Modification: An emerging role in cardiovascular diseases. Journal of Cardiovascular Translational Research, 14, 857–872.
Zhao, W., Li, J., Ma, Q., Cai, J., Li, A., Wu, W., Lv, Y., & Cai, M. (2022). N6-methyladenosine modification participates in neoplastic immunoregulation and tumorigenesis. Journal of Cellular Physiology. https://doi.org/10.1002/jcp.30730
Zhou, M., Liu, W., Zhang, J., & Sun, N. (2021). RNA m(6)A modification in immunocytes and DNA Repair: The biological functions and prospects in clinical application. Frontiers in Cell and Developmental Biology, 9, 794754.
Fu, J., Cui, X., Zhang, X., Cheng, M., Li, X., Guo, Z., & Cui, X. (2021). The role of m6A ribonucleic acid modification in the occurrence of atherosclerosis. Frontiers in Genetics, 12, 733871.
Li, Z., Xu, Q., Huangfu, N., Chen, X., & Zhu, J. (2022). Mettl3 promotes oxLDL-mediated inflammation through activating STAT1 signaling. Journal of Clinical Laboratory Analysis, 36, e24019.
Kumari, R., Dutta, R., Ranjan, P., Suleiman, Z. G., Goswami, S. K., Li, J., Pal, H. C., & Verma, S. K. (2021). ALKBH5 Regulates SPHK1-dependent endothelial cell angiogenesis following ischemic stress. Frontiers in Cardiovascular Medicine, 8, 817304.
Chien, C. S., Li, J. Y., Chien, Y., Wang, M. L., Yarmishyn, A. A., Tsai, P. H., Juan, C. C., Nguyen, P., Cheng, H. M., Huo, T. I., Chiou, S. H., & Chien, S. (2021). METTL3-dependent N(6)-methyladenosine RNA modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proceedings of the National Academy of Sciences of the United States of America. https://doi.org/10.1073/pnas.2025070118
Li, X., Kang, X., Di, Y., Sun, S., Yang, L., Wang, B., & Ji, Z. (2022). CircCHMP5 Contributes to Ox-LDL-induced Endothelial Cell Injury Through the Regulation of MiR-532–5p/ROCK2 axis. Cardiovascular Drugs and Therapy. https://doi.org/10.1007/s10557-022-07316-0
Miao, J., Wang, B., Shao, R., & Wang, Y. (2021). CircUSP36 knockdown alleviates oxidized low-density lipoprotein-induced cell injury and inflammatory responses in human umbilical vein endothelial cells via the miR-20a-5p/ROCK2 axis. International Journal of Molecular Medicine. https://doi.org/10.3892/ijmm.2021.4873
Acknowledgements
No.
Funding
No funding was received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rong, J., Jie, Y. & Zhao, H. m6A ‘writer’ KIAA1429 regulates the proliferation and migration of endothelial cells in atherosclerosis. Mol Biotechnol 65, 1198–1206 (2023). https://doi.org/10.1007/s12033-022-00614-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00614-w