Abstract
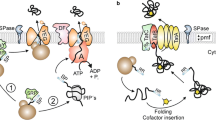
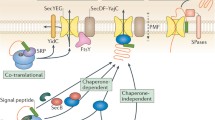
Streptomyces lividans uses mainly two pathways to target secretory proteins to the cytoplasmic membrane. The major pathway (Sec pathway) transports pre-proteins using the signal recognition particle, and the minor Tat pathway is responsible for the secretion using a folded conformation of a relatively low number of proteins. The signal peptides of the Sec-dependent alpha-amylase and the Tat-dependent agarase were interchanged and fused in-frame to the corresponding mature part of the other enzyme. Alpha-amylase was unable to use the Tat route when fused to the agarase signal peptide, while agarase used the Sec route when it was targeted by the alpha-amylase signal peptide. In addition to the signal peptide some yet unidentified parts of the secreted proteins may play a role in selecting the secretory route. Structure predictions for the Tat- and Sec-dependent proteins suggest that less structured proteins are more likely to be candidates for the Tat route.



Similar content being viewed by others
References
Anné, J., Maldonado, B., Van Impe, J., Van Mellaert, L., & Bernaerts, K. (2012). Recombinant protein production and streptomycetes. Journal of Biotechnology, 158, 159–167.
Palacín, A., de la Fuente, R., Valle, I., Rivas, L. A., & Mellado, R. P. (2003). Streptomyces lividans contains a minimal functional signal recognition particle that is involved in protein secretion. Microbiology, 149, 2435–2442.
Berks, B. C., Palmer, T., & Sargent, F. (2005). Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Current Opinion in Microbiology, 8, 174–181.
Jongbloed, J. D., Antelmann, H., Hecker, M., Nijland, R., Bron, S., Airaksinen, U., et al. (2002). Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. Journal of Biological Chemistry, 277, 44068–44078.
Widdick, D. A., Dilks, K., Chandra, G., Bottrill, A., Naldrett, M., Pohlschroder, M., & Palmer, T. (2006). The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proceedings of the National Academy of Sciences of the United States of America, 103, 17927–17932.
Mellado, R. P. (2011). Summing up particular features of protein secretion in Streptomyces lividans. World Journal of Microbiology & Biotechnology, 27, 2231–2237.
Schaerlaekens, K., Schierová, M., Lammertyn, E., Geukens, N., Anné, J., & Van Mellaert, L. (2001). Twin-arginine translocation pathway in Streptomyces lividans. Journal of Bacteriology, 183, 6727–6732.
De Keersmaeker, S., Vrancken, K., Van Mellaert, L., Anné, J., & Geukens, N. (2007). The Tat pathway in Streptomyces lividans: Interaction of Tat subunits and their role in translocation. Microbiology, 153, 1087–1094.
Palmer, T., & Berks, B. C. (2012). The twin-arginine translocation (Tat) protein export pathway. Nature Reviews Microbiology, 10, 483–496.
Hopwood, D. A., Bibb, M. J., & Chater, K. F. (1985). Genetic manipulation of Streptomyces. A laboratory manual. Norwich: John Innes Foundation.
Palomino, C., & Mellado, R. P. (2008). Influence of a Streptomyces lividans SecG functional analogue on protein secretion. International Microbiology, 11, 25–31.
Ward, J. M., Janssen, G. R., Kieser, T., Bibb, M. J., Buttner, M. J., & Bibb, M. J. (1986). Construction and characterisation of a series of multi-copy promoter-probe vectors Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Molecular and General Genetics, 203, 468–475.
Palacín, A., Parro, V., Geukens, N., Anné, J., & Mellado, R. P. (2002). SipY is the Streptomyces lividans type I signal peptidase exerting a major effect on protein secretion. Journal of Bacteriology, 184, 4875–4880.
Parro, V., & Mellado, R. P. (1993). Heterologous recognition in vivo of promoter sequences from the Streptomyces coelicolor dagA gene. FEMS Microbiology Letters, 106, 347–356.
Isiegas, C., Parro, V., & Mellado, R. P. (1999). Streptomyces lividans as a host for the production and secretion of Escherichia coli TEM α-lactamase. Letters in Applied Microbiology, 28, 321–326.
Gust, B., Challis, G. L., Fowler, K., Kieser, T., & Chater, K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterne soil odor geosmin. Proceedings of the National Academy of Sciences of the United States of America, 100, 1541–1546.
Escutia, M. R., Val, G., Palacín, A., Geukens, N., Anné, J., & Mellado, R. P. (2006). Compensatory effect of the minor Streptomyces lividans type I signal peptidases on the SipY major signal peptidase deficiency as determined by extracellular proteome analysis. Proteomics, 6, 4137–4146.
Thompson, C. J., Kieser, T., Ward, J. M., & Hopwood, D. A. (1982). Physical analysis of antibiotic-resistant genes from Streptomyces and their use in vector construction. Gene, 20, 51–62.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Timmons, T. M., & Dunbar, B. S. (1990). Protein blotting and immunodetection. Methods in Enzymology, 182, 679–688.
Gullón, S., Vicente, L. R., & Mellado, R. P. (2012). A novel two-component system involved in secretion stress response in Streptomyces lividans. PLoS ONE, 7(11), e48987. doi:10.1371/journal.pone.0048987.
Parro, V., & Mellado, R. P. (1994). Effect of glucose on agarase overproduction by Streptomyces. Gene, 145, 49–55.
Rabindran, S. K., Haroun, R. I., Wisniewski, J., & Wu, C. (1993). Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science, 259, 230–234.
Rice, P., Longden, I., & Bleasby, A. (2000). EMBOSS: The European molecular biology open software suite. Trends in Genetics, 16, 276–277. doi:10.1016/S0168-9525(00)02024-2.
Cole, C., Barber, J. D., & Barton, G. J. (2008). The Jpred 3 secondary structure prediction server. Nucleics Acids Research, 36(suppl 2), W197–W201.
Prilusky, J., Felder, C. E., Zeev-Ben-Mordehai, T., Rydberg, E. H., Man, O., Beckmann, J. S., et al. (2005). FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics, 21, 3435–3438. doi:10.1093/bioinformatics/bti537.
Lin, H. N., Sung, T. Y., Ho, S. Y., & Hsu, W. L. (2010). Improving protein secondary structure prediction based on short subsequences with local structure similarity. BMC Genomics, 11(Suppl 4), S4. doi:10.1186/1471-2164-11-S4-S4.
Källberg, M., Wang, H., Wang, S., Peng, J., Wang, Z., Lu, H., & Xu, J. (2012). Template-based protein structure modelling using the RaptorX web server. Nature Protocols, 7, 1511–1522. doi:10.1038/nprot.2012.085.
Ferrè, F., & Clote, P. (2006). DIANNA 1.1: An extension of the DIANNA web server for ternary cysteine classification. Nucleic Acids Research, 34(suppl 2), W182–W185.
Schaerlaekens, K., Van Mellaert, L., Lammertyn, E., Geukens, N., & Anné, J. (2004). The importance of the Tat-dependent protein secretion pathway in Streptomyces as revealed by phenotypic changes in tat deletion mutants and genome analysis. Microbiology, 150, 21–31.
Cristóbal, S., de Gier, J. W., Nielsen, H., & von Heijne, G. (1999). Competition between Sec-and TAT-dependent protein translocation in Escherichia coli. EMBO Journal, 18, 2982–2990.
Gullón, S., Palomino, C., Navajas, R., Paradela, A., & Mellado, R. P. (2012). Translocase and major signal peptidase malfunctions affect aerial mycelium formation in Streptomyces lividans. Journal of Biotechnology, 160, 112–122.
Acknowledgments
The Spanish Ministry of the Environment and Rural and Marine affairs has commissioned and supported this research (Grant No. EGO22008). Grant PIE201220E003 from the CSIC also supported this research. We wish to thank C. Palomino for her contribution to obtaining S. lividans ∆tatC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gullón, S., Vicente, R.L., Valverde, J.R. et al. Exploring the Feasibility of the Sec Route to Secrete Proteins Using the Tat Route in Streptomyces lividans . Mol Biotechnol 57, 931–938 (2015). https://doi.org/10.1007/s12033-015-9883-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9883-0