Abstract
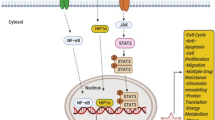
Triple-negative breast cancers (TNBCs) have poor prognosis, and chemotherapy remains the mainstay of therapy because of lack of discovered possible target. MYC were found overexpressed in TNBCs compared with other subtypes and especially in those resistant to chemotherapy, but the inhibition has been challenging to achieve. Recently, the cooperation of PIM1 and MYC was identified involved in cell proliferation, migration and apoptosis of TNBCs, which has been reported in hematological malignancy and prostatic cancer. Inhibition of PIM1 can promote the apoptosis of tumor cells and enhance sensitivity to chemotherapy. Notably, PIM1-null mice develop normally and are fertile, suggesting the side effects can be tolerated. Thus, PIM1 may be a promising target in TNBCs and further investigation, both in vivo and in vitro, needs to be carried out.



Similar content being viewed by others
References
DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. doi:10.3322/caac.21203.
Newman LA, Reis-Filho JS, Morrow M, Carey LA, King TA. The 2014 Society of Surgical Oncology Susan G. Komen for the Cure Symposium: triple-negative breast cancer. Ann Surg Oncol. 2015;22(3):874–82. doi:10.1245/s10434-014-4279-0.
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 Pt 1):4429–34. doi:10.1158/1078-0432.ccr-06-3045.
Peto R, Davies C, Godwin J, Gray R, Pan HC, Clarke M, et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet (Lond, Engl). 2012;379(9814):432–44. doi:10.1016/s0140-6736(11)61625-5.
Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet (Lond, Engl). 2014;384(9938):164–72. doi:10.1016/s0140-6736(13)62422-8.
Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4(2):232–45. doi:10.1158/2159-8290.cd-13-0286.
Wang J, Kim J, Roh M, Franco OE, Hayward SW, Wills ML, et al. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene. 2010;29(17):2477–87. doi:10.1038/onc.2010.10.
Keeton EK, McEachern K, Dillman KS, Palakurthi S, Cao Y, Grondine MR, et al. AZD1208, a potent and selective pan-Pim kinase inhibitor, demonstrates efficacy in preclinical models of acute myeloid leukemia. Blood. 2014;123(6):905–13. doi:10.1182/blood-2013-04-495366.
Braso-Maristany F, Filosto S, Catchpole S, Marlow R, Quist J, Francesch-Domenech E, et al. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med. 2016;22(11):1303–13. doi:10.1038/nm.4198.
Horiuchi D, Camarda R, Zhou AY, Yau C, Momcilovic O, Balakrishnan S, et al. PIM1 kinase inhibition as a targeted therapy against triple-negative breast tumors with elevated MYC expression. Nat Med. 2016;22(11):1321–9. doi:10.1038/nm.4213.
Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22(20):2755–66. doi:10.1101/gad.1712408.
Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17(9):1115–29. doi:10.1101/gad.1067003.
Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19(1):1–11.
Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–90. doi:10.1038/nrc2231.
Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–45. doi:10.1016/j.cell.2010.03.030.
Jain AN, Chin K, Borresen-Dale AL, Erikstein BK, Eynstein Lonning P, Kaaresen R, et al. Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival. Proc Natl Acad Sci USA. 2001;98(14):7952–7. doi:10.1073/pnas.151241198.
Horiuchi D, Kusdra L, Huskey NE, Chandriani S, Lenburg ME, Gonzalez-Angulo AM, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med. 2012;209(4):679–96. doi:10.1084/jem.20111512.
Chandriani S, Frengen E, Cowling VH, Pendergrass SA, Perou CM, Whitfield ML, et al. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS ONE. 2009;4(8):e6693. doi:10.1371/journal.pone.0006693.
Alles MC, Gardiner-Garden M, Nott DJ, Wang Y, Foekens JA, Sutherland RL, et al. Meta-analysis and gene set enrichment relative to er status reveal elevated activity of MYC and E2F in the “basal” breast cancer subgroup. PLoS ONE. 2009;4(3):e4710. doi:10.1371/journal.pone.0004710.
Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci USA. 2010;107(15):6994–9. doi:10.1073/pnas.0912708107.
Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2(4):333–44. doi:10.1016/j.stem.2008.02.009.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi:10.1038/ng.127.
Chang DW, Claassen GF, Hann SR, Cole MD. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol Cell Biol. 2000;20(12):4309–19.
Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14(19):2501–14.
Seo HR, Kim J, Bae S, Soh JW, Lee YS. Cdk5-mediated phosphorylation of c-Myc on Ser-62 is essential in transcriptional activation of cyclin B1 by cyclin G1. J Biol Chem. 2008;283(23):15601–10. doi:10.1074/jbc.M800987200.
Prochownik EV, Vogt PK. Therapeutic targeting of Myc. Genes Cancer. 2010;1(6):650–9. doi:10.1177/1947601910377494.
Cuypers HT, Selten G, Quint W, Zijlstra M, Maandag ER, Boelens W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37(1):141–50.
van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56(4):673–82.
Narlik-Grassow M, Blanco-Aparicio C, Carnero A. The PIM family of serine/threonine kinases in cancer. Med Res Rev. 2014;34(1):136–59. doi:10.1002/med.21284.
Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 2010;95(6):1004–15. doi:10.3324/haematol.2009.017079.
Saris CJ, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991;10(3):655–64.
Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosom Cancer. 2006;45(11):1033–40. doi:10.1002/gcc.20366.
Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15(2):441–51. doi:10.1158/1078-0432.ccr-08-1791.
Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29(14):2013–23. doi:10.1038/onc.2009.489.
Bazarov AV, Hines WC, Mukhopadhyay R, Beliveau A, Melodyev S, Zaslavsky Y, et al. Telomerase activation by c-Myc in human mammary epithelial cells requires additional genomic changes. Cell Cycle (Georgetown, TX). 2009;8(20):3373–8. doi:10.4161/cc.8.20.9856.
Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9(8):932–44. doi:10.1038/ncb1618.
Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17(15):1841–54. doi:10.1101/gad.1105003.
Yan B, Zemskova M, Holder S, Chin V, Kraft A, Koskinen PJ, et al. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem. 2003;278(46):45358–67. doi:10.1074/jbc.M307933200.
Dautry F, Weil D, Yu J, Dautry-Varsat A. Regulation of pim and myb mRNA accumulation by interleukin 2 and interleukin 3 in murine hematopoietic cell lines. J Biol Chem. 1988;263(33):17615–20.
Mochizuki T, Kitanaka C, Noguchi K, Muramatsu T, Asai A, Kuchino Y. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J Biol Chem. 1999;274(26):18659–66.
Bachmann M, Kosan C, Xing PX, Montenarh M, Hoffmann I, Moroy T. The oncogenic serine/threonine kinase Pim-1 directly phosphorylates and activates the G2/M specific phosphatase Cdc25C. Int J Biochem Cell Biol. 2006;38(3):430–43. doi:10.1016/j.biocel.2005.10.010.
Wang Z, Bhattacharya N, Mixter PF, Wei W, Sedivy J, Magnuson NS. Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochem Biophys Acta. 2002;1593(1):45–55.
Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68(13):5076–85. doi:10.1158/0008-5472.can-08-0634.
Zhang Y, Wang Z, Magnuson NS. Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol Cancer Res. 2007;5(9):909–22. doi:10.1158/1541-7786.mcr-06-0388.
Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571(1–3):43–9. doi:10.1016/j.febslet.2004.06.050.
Chao DT, Linette GP, Boise LH, White LS, Thompson CB, Korsmeyer SJ. Bcl-XL and Bcl-2 repress a common pathway of cell death. J Exp Med. 1995;182(3):821–8.
Bachmann M, Moroy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005;37(4):726–30. doi:10.1016/j.biocel.2004.11.005.
Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Investig. 2005;115(10):2618–24. doi:10.1172/jci26273.
Wang Z, Bhattacharya N, Weaver M, Petersen K, Meyer M, Gapter L, et al. Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J Vet Sci. 2001;2(3):167–79.
White E. The pims and outs of survival signaling: role for the Pim-2 protein kinase in the suppression of apoptosis by cytokines. Genes Dev. 2003;17(15):1813–6. doi:10.1101/gad.1123103.
Yip-Schneider MT, Horie M, Broxmeyer HE. Transcriptional induction of pim-1 protein kinase gene expression by interferon gamma and posttranscriptional effects on costimulation with steel factor. Blood. 1995;85(12):3494–502.
Matikainen S, Sareneva T, Ronni T, Lehtonen A, Koskinen PJ, Julkunen I. Interferon-alpha activates multiple STAT proteins and upregulates proliferation-associated IL-2Ralpha, c-myc, and pim-1 genes in human T cells. Blood. 1999;93(6):1980–91.
Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011;11(1):23–34. doi:10.1038/nrc2986.
Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, et al. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24(13):6104–15. doi:10.1128/mcb.24.13.6104-6115.2004.
Block KM, Hanke NT, Maine EA, Baker AF. IL-6 stimulates STAT3 and Pim-1 kinase in pancreatic cancer cell lines. Pancreas. 2012;41(5):773–81. doi:10.1097/MPA.0b013e31823cdd10.
Mizuno K, Shirogane T, Shinohara A, Iwamatsu A, Hibi M, Hirano T. Regulation of Pim-1 by Hsp90. Biochem Biophys Res Commun. 2001;281(3):663–9. doi:10.1006/bbrc.2001.4405.
Shay KP, Wang Z, Xing PX, McKenzie IF, Magnuson NS. Pim-1 kinase stability is regulated by heat shock proteins and the ubiquitin-proteasome pathway. Mol Cancer Res. 2005;3(3):170–81. doi:10.1158/1541-7786.mcr-04-0192.
Malinen M, Jaaskelainen T, Pelkonen M, Heikkinen S, Vaisanen S, Kosma VM, et al. Proto-oncogene PIM-1 is a novel estrogen receptor target associating with high grade breast tumors. Mol Cell Endocrinol. 2013;365(2):270–6. doi:10.1016/j.mce.2012.10.028.
Laird PW, van der Lugt NM, Clarke A, Domen J, Linders K, McWhir J, et al. In vivo analysis of Pim-1 deficiency. Nucleic Acids Res. 1993;21(20):4750–5.
Abadi AH, Abouel-Ella DA, Lehmann J, Tinsley HN, Gary BD, Piazza GA, et al. Discovery of colon tumor cell growth inhibitory agents through a combinatorial approach. Eur J Med Chem. 2010;45(1):90–7. doi:10.1016/j.ejmech.2009.09.029.
Magnuson NS, Wang Z, Ding G, Reeves R. Why target PIM1 for cancer diagnosis and treatment? Future Oncol. 2010;6(9):1461–78. doi:10.2217/fon.10.106.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The studies with human participants or animals involved in our manuscript have been listed in references, and our team did not perform any studies with human participants or animals about PIM1 and corresponding molecules.
Rights and permissions
About this article
Cite this article
Zhao, W., Qiu, R., Li, P. et al. PIM1: a promising target in patients with triple-negative breast cancer. Med Oncol 34, 142 (2017). https://doi.org/10.1007/s12032-017-0998-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-017-0998-y