Abstract
Published data on the association between DNA methyltransferase (DNMT) 3B −149C/T polymorphism and cancer risk remain inconclusive. To derive a more precise estimation for this association, we performed a meta-analysis of 5,903 cancer cases and 8,132 controls from 22 published case–control studies. We used odds ratios (ORs) with 95 % confidence intervals (CIs) to assess the strength of the association. Our meta-analysis suggested that DNMT3B −149C/T polymorphism was associated with the risk of head and neck cancer under heterozygote comparison (OR 0.73, 95 % CI 0.59–0.90) and dominant model (OR 1.75, 95 % CI 0.62–0.92), although no evidence of association between DNMT3B −149C/T polymorphism and cancer risk was observed as we compared in the pooled analyses (homozygote comparison: OR 0.96, 95 % CI 0.86–1.09; heterozygote comparison: OR 1.07, 95 % CI 0.86–0.32; dominant model: OR 1.03, 95 % CI 0.85–1.25; recessive model: OR 0.93, 95 % CI 0.8–1.08). More studies are needed to detect DNMT3B −149C/T polymorphism and its association with cancer in different ethnic populations incorporated with environment exposures in the susceptibility of different kinds of cancer.
Similar content being viewed by others
Introduction
DNA methylation is a major epigenetic modification that involves the addition of a methyl group to the 5′ position of a cytosine in a CpG dinucleotide, which is catalyzed by a family of DNA methyltransferases (DNMTs) including three activated forms (DNMT1, DNMT3A and DNMT3B) in human [1, 2]. Aberrant DNA methylation is one of the most consistent epigenetic changes observed in human cancers [3]. DNMT1 is a maintenance DNA methyltransferase, whereas DNMT3A and DNMT3B are considered as de novo methyltransferases because they can methylate unmethylated and hemi-methylated DNA with equal efficiency [4, 5]. A number of studies showed that DNMT3B was up-regulated in several human cancers, demonstrating that DNMT3B may play an important role in tumorigenesis by contributing to the generation of aberrant DNA methylation [6–8].
The DNMT3B gene is assigned to chromosome 20q11.2 and contains a single C→T transition polymorphism (C46359T) at a novel promoter region, −149 base pairs from the transcription start site, which may result in greatly increased promoter activity of the gene [9]. A number of single-nucleotide polymorphisms (SNP) in the gene have been described in the literature, of which a common SNP −149C/T (rs2424913) in the promoter region of the DNMT3B is known to regulate its expression [10]. Recently, a variety of molecular epidemiological studies have been conducted to examine the association between DNMT3B −149C/T polymorphism and cancer susceptibility [11–31], but the results remain inconclusive. Therefore, the association between DNMT3B −149C/T polymorphism and cancer risk requires further investigation.
Considering the relatively small sample size in most studies, it is possible to perform a quantitative synthesis of the evidence with rigorous methods. Here, we performed a meta-analysis on 22 published case–controls to derive a more precise evaluation of the association between DNMT3B −149C/T polymorphism and cancer risk.
Materials and methods
Identification and eligibility of relevant studies
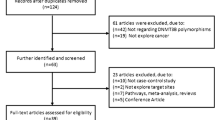
A systematic literature search was performed using PubMed, Medline, EMBASE and Chinese National Knowledge Infrastructure (CNKI), covering all articles published up to October 2014. We used the following terms: “DNMT3B,” “polymorphism,” “rs2424913” and “cancer”. References of the retrieved publications were also screened. All eligible studies were retrieved, and their bibliographies were checked for other relevant publications. Only published studies with full-text articles were included. When overlapping articles were found, we only included the publications that reported the most extensive information.
Inclusion criteria
The inclusion criteria were as follows: (1) published in English or in Chinese; (2) case–control studies of cancer with DNMT3B −149C/T polymorphism; (3) supply the available genotype frequencies in cancer cases and controls; and (4) sufficient published data for estimating an odds ratio (OR) with 95 % confidence interval (CI).
Data extraction
Two investigators independently (Jing Zhu and Songtao Du) reviewed the articles to exclude irrelevant and overlapping studies. The results were compared, and disagreements were resolved by discussion and consensus. We extracted the following information from each study: first author’s surname, year, ethnicity, tumor type, definition of cases, characteristics of controls, validity of the genotyping method, and the number of cases and controls for each genotype.
Statistical analysis
OR and 95 % CI were used to assess the strength of association between DNMT3B −149C/T polymorphism and the risk of cancer under homozygote comparison (CC vs. TT), heterozygote comparison (CT vs. TT), dominant (CC/CT vs. TT) and recessive (CC vs. CT/TT) genetic model comparison. The significance of the combined OR was determined by the Z test, in which P < 0.05 was considered significant. Stratified analyses were also performed by cancer types, ethnicities, and sources of controls. The Chi-square-based Q statistic test was performed to evaluate the between-study heterogeneity of studies. If P < 0.1, between-study heterogeneity was considered to be significant [32]. When the effects were assumed to be homogenous, the fixed effects model based on Peto method was used, otherwise, the random effects model based on Mantel–Haenszel method was applied. We also used the statistic of I 2 to efficiently test for the heterogeneity, with I 2 < 25 %, 25–75 % and >75 % to represent low, moderate and high degree of inconsistency, respectively [33]. Funnel plots were used to access the potential publication bias by the method of Egger’s linear regression test [34]. All analyses were performed by Stata (version 10.0, Stata Corporation) and Review Manager (version 5.0.0, The Cochrane collaboration), using two side P values.
Results
Characteristics of studies
Twenty two case–control studies including 5,903 cancer cases and 8,132 controls met the including criteria. The study characteristics were listed in Table 1. Most of cases in the studies were histologically diagnosed, and most of the controls were selected from healthy population. Fifteen studies used frequency-matched controls to the cases by age, sex, residence or ethnicity. A classic polymerase chain reaction–restriction fragment length polymorphism assay was performed in all studies (Table 1).
Main results
The evaluation of association between DNMT3B −149C/T polymorphism and cancer risk is presented in Table 2. There was no significant association between DNMT −149C/T polymorphism and the risk of cancer (CC vs. TT: OR 0.96, 95 % CI 0.86–1.09; P = 0.1, I 2 = 34 % for heterogeneity). In the stratified analysis by cancer type, DNMT3B −149C/T polymorphism was relative with a significantly increased risk of head and neck cancer in two tested models (CT vs. TT: OR 0.73, 95 % CI 0.59–0.9; P = 0.33, I 2 = 0 % for heterogeneity; CC/CT vs. TT: OR 0.76, 95 % CI 0.61–0.93; P = 0.3, I 2 = 7 % for heterogeneity; Fig. 1). However, no significant elevated risk of colorectal cancer, gastric cancer, hepatocellular cancer, breast cancer and other cancers with this polymorphism were shown in overall comparisons. At the same time, we failed to find significant main effects for DNMT3B −149C/T polymorphism on cancer risk in different genetic models when stratified according to ethnicity or sources of controls.
Test of heterogeneity
There was significant heterogeneity for recessive model comparison (CC vs. CT/TT: P heterogeneity = 0.01), for heterozygote comparison (CT vs. TT: P heterogeneity < 0.001) and for dominant model comparison (CC/CT vs. TT: P heterogeneity < 0.001), but not for homozygote comparison and (CC/TT: P heterogeneity = 0.1). Then, we assessed the source of heterogeneity for homozygote comparison by cancer type, ethnicity and source of controls. As a result, cancer type (χ 2 = 7.04, df = 4, P = 0.13), ethnicity (χ 2 = 3.36, df = 3, P = 0.34) and source of controls (χ 2 = 2.56, df = 1, P = 0.11) were not found to contribute to substantial heterogeneity.
Sensitivity analysis
Sensitivity analysis was performed by sequential omission of individual studies in whole subjects and subgroups, respectively. For DNMT3B −149C/T, the significance of pooled ORs was influenced evidently by individual study on the whole population or subgroup analysis of cancer type and ethnicity. In the cancer type subgroup analysis, the study of Jones et al. [13] was the main originators of heterogeneity in the colorectal cancer. When the study was excluded, heterogeneity was significantly decreased (CC vs. CT/TT: P heterogeneity = 0.94, I 2 = 0 %). Similarly, when study by Mostowska et al. [31] was excluded, heterogeneity was also decreased in other type cancer (CC vs. CT/TT: P heterogeneity = 0.34, I 2 = 11 %). Additionally, in the ethnicity subgroup analysis, sensitivity analyses suggested that the study [28] was the main originator of heterogeneity in Asian. After exclusion of this study, heterogeneity was significantly decreased (CT vs. TT: Pheterogeneity = 0.37, I 2 = 8 %; CC/CT vs. TT: Pheterogeneity = 0.37, I 2 = 0 %).
Publication bias
Funnel plots are shown in Fig. 2 for dominant model. Arrangement of data points did not reveal any evidence of obvious asymmetry. Formal evaluation using Egger’s regression asymmetry tests for dominant model and the result still did not show any evidence of publication bias (t = 0.25, P = 0.80).
Discussion
The present meta-analysis, including 5,903 cancer cases and 8,132 controls from 22 published case–control studies, showed that the DNMT3B −149C/T was not associated with cancer risk. When stratified by different types of cancer, we found an association between DNMT3B −149C/T polymorphism and head and neck cancer risk under heterozygote comparison and dominant model, but there are only two studies in analysis with limited sample size; therefore, the result should be interpreted with caution. Given the important roles of DNMT3B in cancer risk, it was biologically possible that DNMT3B polymorphism is associated with the risk of cancer by increasing DNMT3B promoter activity that modulated an aberrant de novo methylation of CpG islands in some tumor suppressor genes [4]. Studies on the functionality of this polymorphism might contribute to a better understanding of tumor biology and behavior and help us to predict the genetic susceptibility of cancer and choose therapies in an individual manner. However, DNMT3B −149C/T polymorphism did not increase the risk of colorectal cancer, gastric cancer, breast cancer and hepatocellular carcinoma in overall population. The probability may be that different types of cancer may have different mechanism of carcinogenesis. The differences in genetic background and/or environmental exposure may result in different frequency of −149 C/T genotype in healthy individuals from distinct ethnicities; however, in subgroup analysis by ethnicity, we also did not find that DNMT3B −149C/T was associated with ethnicity. It is likely that the small sample size may have insufficient statistical power to detect a real effect. Therefore, more studies based on large population and more different ethnicity should be conducted to further examine this association.
Heterogeneity is a potential problem when interpreting the results of all meta-analysis. Although we minimized the likelihood by performing a careful search for published studies, using strict criteria for study inclusion, precise data extraction and careful data analysis, significant between-study heterogeneity existed in most comparisons. After subgroup analysis by cancer types, ethnicity and source of controls, the heterogeneity was effectively decreased, but significant heterogeneity still existed. Thus, we choose to use random effects model, when I 2 value for heterogeneity test is <50 %. The reason might be that different genetic backgrounds and the environment existed among different ethnicities and individuals.
Numbers of SNPs, however, were frequently investigated in the former studies to evaluate the association between DNMT3B polymorphisms and cancer in diverse populations. There might be some other SNPs in DNMT3B associated with risk of cancer. Lee et al. [35] found C alleles of DNMT3B contributed to the susceptibility of lung cancer in Korean population. Some other SNPs of DNMT3B, such as −579 G/T and −283 T/C, were also researched by some studies on their association with cancer risk [11, 12, 14, 19, 36, 37]. However, there were only a very limited number of studies available for some SNPs and therefore not having enough statistical power to explore the real association.
Some other limitations in our meta-analysis should be acknowledged. Firstly, controls were not uniformly defined, while our result was based on unadjusted estimates. Secondly, in the subgroup analyses, the sample size of different types of cancer was relatively small, such as lung cancer, ovarian cancer and prostate cancer not having enough statistical power to explore the real association. Thirdly, only English and Chinese language studies were included in this meta-analysis might have led to publication bias, and the exclusion of unpublished data was generally associated with an overestimation of the true effect.
In conclusion, our meta-analysis suggested that DNMT3B −149C/T polymorphism was not related to overall cancer risk, whereas there was an association between DNMT3B −149C/T polymorphism and head and neck cancer risk under heterozygote comparison and dominant model. Larger samples among different populations, especially more sophisticated gene–gene and gene–environment interactions should be considered in future studies, which should lead to better, comprehensive understanding of the association between DNMT3B −149C/T polymorphism and cancer risk.
References
Bheemanaik S, Reddy YV, Rao DN. Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem J. 2006;399:177–90.
Xie S, et al. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95.
Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–7.
Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57.
Robertson KD, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and over expression in tumors. Nucleic Acids Res. 1999;27:2291–8.
Belinsky SA, Nikula KJ, Baylin SB, Issa PJ. Increased cytosine DNA methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci. 1996;93:4045–50.
Nagai M, Nakamura A, Makino R, Mitamura K. Expression of DNA (5-cytosin)-methyltransferases (DNMTs) in hepatocellular carcinomas. Hepatol Res. 2003;26:186–91.
Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108–13.
Wang L, et al. Polymorphism in DNMT3B6 promoter region and lung cancer risk. Proc Am Assoc Cancer Res. 2001;42:863.
Shen H, et al. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002;62:4992–5.
Bao Q, et al. Genetic variation in the promoter of DNMT3B is associated with the risk of colorectal cancer. Int J Colorectal Dis. 2011;9:1107–12.
Fan H, Zhang F, Hu J, Liu D, Zhao Z. Promoter polymorphisms of DNMT3B and the risk of colorectal cancer in Chinese: a case–control study. J Exp Clin Cancer Res. 2008;27:24.
Jones JS, et al. DNMT3b polymorphism and hereditary nonpolyposis colorectal cancer age of onset. Cancer Epidemiol Biomark Prev. 2006;15:886–91.
Karpinski P, et al. Polymorphisms in methyl-group metabolism genes and risk of sporadic colorectal cancer with relation to the CpG island methylator phenotype. Cancer Epidemiol. 2010;34:338–44.
de Vogel S, et al. Genetic variants of methyl metabolizing enzymes and epigenetic regulators: associations with promoter CpG Island hypermethylation in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3086–96.
Iacopetta B, et al. The MTHFR C677T and ΔDNMT3B C −149T polymorphisms confer different risks for right and left-sided colorectal cancer. Int J Cancer. 2009;125:84–90.
Reeves SG, et al. The −149C>T SNP within the ΔDNMT3B gene, is not associated with early disease onset in hereditary non-polyposis colorectal cancer. Cancer Lett. 2008;265:39–44.
Aung PP, et al. No evidence of correlation between the single nucleotide polymorphism of DNMT3B promoter and gastric cancer risk in a Japanese population. Oncol Rep. 2005;14:1151–4.
Hu J, et al. DNMT3B promoter polymorphism and risk of gastric cancer. Diq Dis Sci. 2010;55:1011–6.
Wang YM, et al. Single nucleotide polymorphism in DNA methyltransferase 3B promoter and its association with gastric cardiac adenocarcinoma in North China. World J Gastroenterol. 2005;11:3623–7.
Succi M, et al. DNMT3B C46359T and SHMT1 C1420T polymorphisms in the folate pathway in carcinogenesis of head and neck. Mol Biol Rep. 2013;41:581–9.
Liu Z, Wang L, Wang LE, Sturqis EM, Wei Q. Polymorphisms of the DNMT3B gene and risk of squamous cell carcinoma of the head and neck: a case–control study. Cancer Lett. 2008;268:158–65.
Ezzikouri S, et al. Single nucleotide polymorphism in DNMT3B promoter and its association with hepatocellular carcinoma in a Moroccan population. Infect Genet Evol. 2009;9:877–81.
Wu Y, Lin JS. DNA methyltransferase 3B promoter polymorphism and its susceptibility to primary hepatocellular carcinoma in the Chinese Han nationality population: a case–control study. World J Gastroenterol. 2007;13:6082–6.
Lao Y, et al. Promoter polymorphisms of DNA methyltransferase 3B and risk of hepatocellular carcinoma. Biomed Rep. 2013;1:771–5.
Eftekhar E, Rasti M, Nahgibalhossaini F, Sadeghi Y. The study of DNA methyltransferase-3B promoter variant genotype among Iranian. Iran J Med Sci. 2014;3:268–74.
Montgomery KG, Liu MC, Eccles DM, Campbell IG. The DNMT3B C→T promoter polymorphism and risk of breast cancer in a British population: a case–control study. Breast Cancer Res. 2004;6:R390–4.
Li Y, et al. The C46359T polymorphism of DNMT3B promoter gene and pathogenesis of acute leukemia. Zhonghua Nei Ke Za Zhi. 2005;44:588–91.
Singal R, Das PM, Manoharan M, Reis IM, Schlesselman JJ. Polymorphisms in the DNA methyltransferase 3b gene and prostate cancer risk. Oncol Rep. 2005;14:569–73.
Hernández-Sotelo D, et al. The 46359CT polymorphism of DNMT3B is associated with the risk of cervical cancer. Mol Biol Rep. 2013;7:4275–80.
Mostowska A, Sajdak S, Pawlik P, Lianeri M. Jagodzinski PP. DNMT1, DNMT3A and DNMT3B gene variants in relation to ovarian cancer risk in the Polish population. Mol Biol Rep. 2013;40:4893–9.
Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol. 2005;28:123–37.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60.
Egger M. Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34.
Lee SJ, et al. DNMT3B polymorphism sandrisk of primary lung cancer. Carcinogenesis. 2005;26:403–9.
Chang KP, et al. Promoter polymorphisms of DNMT3B and the risk of head and neck squamous cell carcinoma in Taiwan: a case–control study. Oral Oncol. 2007;43:345–51.
Chang KP, et al. Gene expression and promoter polymorphisms of DNA methyltransferase 3B in nasopharyngeal carcinomas in Taiwanese people: a case–control study. Oncol Rep. 2008;19:217–22.
Conflict of interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Zhu, J., Du, S., Zhang, J. et al. Polymorphism of DNA methyltransferase 3B −149C/T and cancer risk: a meta-analysis. Med Oncol 32, 399 (2015). https://doi.org/10.1007/s12032-014-0399-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-014-0399-4