Abstract
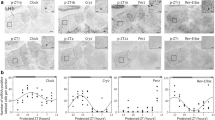
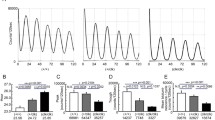
Early growth response transcription factor 1 (EGR1) is expressed in the suprachiasmatic nucleus (SCN) after light stimulation. We used EGR1-deficient mice to address the role of EGR1 in the clock function and light-induced resetting of the clock. The diurnal rhythms of expression of the clock genes BMAL1 and PER1 in the SCN were evaluated by semi-quantitative in situ hybridization. We found no difference in the expression of PER1 mRNA between wildtype and EGR1–deficient mice; however, the daily rhythm of BMAL1 mRNA was completely abolished in the EGR1–deficient mice. In addition, we evaluated the circadian running wheel activity, telemetric locomotor activity, and core body temperature of the mice. Loss of EGR1 neither altered light-induced phase shifts at subjective night nor affected negative masking. Overall, circadian light entrainment was found in EGR1-deficient mice but they displayed a reduced locomotor activity and an altered temperature regulation compared to wild type mice. When placed in running wheels, a subpopulation of EGR1-deficient mice displayed a more disrupted activity rhythm with no measurable endogenous period length (tau). In conclusion, the present study provides the first evidence that the circadian clock in the SCN is disturbed in mice deficient of EGR1.





Similar content being viewed by others
References
Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S (1999) Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci 19:1115–1121
Albrecht U, Sun ZS, Eichele G, Lee CC (1997) A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 91:1055–1064
Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC (2001) MPer1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythm 16:100–104. https://doi.org/10.1177/074873001129001791
Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR (2001) Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30:525–536
Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009–1017
Dong YN, Goguen D, Robertson HA, Rusak B (2002) Anatomical and temporal differences in the regulation of ZIF268 (NGFI-A) protein in the hamster and mouse suprachiasmatic nucleus. Neuroscience 111:567–574
Daan S, Pittendrigh CS (1976) A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol 106:253–266. https://doi.org/10.1007/bf01417857
Golombek DA, Rosenstein RE (2010) Physiology of circadian entrainment. Physiol Rev 90:1063–1102. https://doi.org/10.1152/physrev.00009.2009
Hannibal J, Brabet P, Fahrenkrug J (2008) Mice lacking the PACAP type I receptor have impaired photic entrainment and negative masking. Am J Physiol Regul Integr Comp Physiol 295:R2050–R2058. https://doi.org/10.1152/ajpregu.90563.2008
Hannibal J, Hsiung HM, Fahrenkrug J (2011) Temporal phasing of locomotor activity, heart rate rhythmicity, and core body temperature is disrupted in VIP receptor 2-deficient mice. Am J Physiol Regul Integr Comp Physiol 300:R519–R530. https://doi.org/10.1152/ajpregu.00599.2010
Hannibal J, Jamen F, Nielsen HS, Journot L, Brabet P, Fahrenkrug J (2001) Dissociation between light-induced phase shift of the circadian rhythm and clock gene expression in mice lacking the pituitary adenylate cyclase activating polypeptide type 1 receptor. J Neurosci 21:4883–4890
Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH (2002) The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109:497–508
Honrado GI, Johnson RS, Golombek DA, Spiegelman BM, Papaioannou VE, Ralph MR (1996) The circadian system of c-fos deficient mice. J Comp Physiol A 178:563–570
Hughes AT, Piggins HD (2008) Behavioral responses of Vipr2−/− mice to light. J Biol Rhythm 23:211–219. https://doi.org/10.1177/0748730408316290
Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S (2001) A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci 4:289–296. https://doi.org/10.1038/85138
Kilduff TS, Vugrinic C, Lee SL, Milbrandt JD, Mikkelsen JD, O'Hara BF, Heller HC (1998) Characterization of the circadian system of NGFI-A and NGFI-A/NGFI-B deficient mice. J Biol Rhythm 13:347–357. https://doi.org/10.1177/074873098129000174
Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS (1990) Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron 5:127–134
Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS (1992) Regulation of jun-B messenger RNA and AP-1 activity by light and a circadian clock. Science 255:1581–1584
Kuroda H, Fukushima M, Nakai M, Katayama T, Murakami N (1997) Daily wheel running activity modifies the period of free-running rhythm in rats via intergeniculate leaflet. Physiol Behav 61:633–637
Lee JL, Everitt BJ, Thomas KL (2004) Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science 304:839–843. https://doi.org/10.1126/science.1095760
Li L, Carter J, Gao X, Whitehead J, Tourtellotte WG (2005) The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol Cell Biol 25:10286–10300. https://doi.org/10.1128/mcb.25.23.10286-10300.2005
Lin JT, Kornhauser JM, Singh NP, Mayo KE, Takahashi JS (1997) Visual sensitivities of nur77 (NGFI-B) and zif268 (NGFI-A) induction in the suprachiasmatic nucleus are dissociated from c-fos induction and behavioral phase-shifting responses. Brain Res Mol Brain Res 46:303–310
Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, Zhang AW, Parcy F, Lenhard B, Sandelin A, Wasserman WW (2016) JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44:D110–D115. https://doi.org/10.1093/nar/gkv1176
McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, Takahashi JS (2006) Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 314:1304–1308. https://doi.org/10.1126/science.1132430
Mohawk JA, Green CB, Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35:445–462. https://doi.org/10.1146/annurev-neuro-060909-153128
Mohawk JA, Takahashi JS (2011) Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends Neurosci 34:349–358. https://doi.org/10.1016/j.tins.2011.05.003
Nelson W, Tong YL, Lee JK, Halberg F (1979) Methods for cosinor-rhythmometry. Chronobiologia 6:305–323
Nielsen HS, Hannibal J, Knudsen SM, Fahrenkrug J (2001) Pituitary adenylate cyclase-activating polypeptide induces period1 and period2 gene expression in the rat suprachiasmatic nucleus during late night. Neuroscience 103:433–441
Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U (2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251–260
Resuehr HE, Resuehr D, Olcese J (2009) Induction of mPer1 expression by GnRH in pituitary gonadotrope cells involves EGR-1. Mol Cell Endocrinol 311:120–125. https://doi.org/10.1016/j.mce.2009.07.005
Rusak B, McNaughton L, Robertson HA, Hunt SP (1992) Circadian variation in photic regulation of immediate-early gene mRNAs in rat suprachiasmatic nucleus cells. Brain Res Mol Brain Res 14:124–130
Rusak B, Robertson HA, Wisden W, Hunt SP (1990) Light pulses that shift rhythms induce gene expression in the suprachiasmatic nucleus. Science 248:1237–1240
Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR (2000a) Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol 20:6269–6275
Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM (2000b) Interacting molecular loops in the mammalian circadian clock. Science 288:1013–1019
Shi S, Hida A, McGuinness OP, Wasserman DH, Yamazaki S, Johnson CH (2010) Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr Biol 20:316–321. https://doi.org/10.1016/j.cub.2009.12.034
Swirnoff AH, Milbrandt J (1995) DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol 15:2275–2287
Tao W, Wu J, Zhang Q, Lai SS, Jiang S, Jiang C, Xu Y, Xue B, Du J, Li CJ (2015) EGR1 regulates hepatic clock gene amplitude by activating Per1 transcription. Sci Rep 5:15212. https://doi.org/10.1038/srep15212
Wei F, Xu ZC, Qu Z, Milbrandt J, Zhuo M (2000) Role of EGR1 in hippocampal synaptic enhancement induced by tetanic stimulation and amputation. J Cell Biol 149:1325–1334
Yu W, Nomura M, Ikeda M (2002) Interactivating feedback loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2. Biochem Biophys Res Commun 290:933–941. https://doi.org/10.1006/bbrc.2001.6300
Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC (2001) Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105:683–694
Zylka MJ, Shearman LP, Weaver DR, Reppert SM (1998) Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20:1103–1110
Acknowledgements
The skillful technical assistance of Anita Hansen and Tina Wintersø is gratefully acknowledged. The study was supported by the Danish Biotechnology Center for Cellular Communication.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Riedel, C.S., Georg, B., Jørgensen, H.L. et al. Mice Lacking EGR1 Have Impaired Clock Gene (BMAL1) Oscillation, Locomotor Activity, and Body Temperature. J Mol Neurosci 64, 9–19 (2018). https://doi.org/10.1007/s12031-017-0996-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-017-0996-8