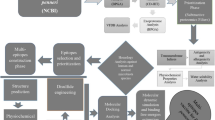
Abstract
Brucella suis mediates the transmission of brucellosis in humans and animals and a significant facultative zoonotic pathogen found in livestock. It has the capacity to survive and multiply in a phagocytic environment and to acquire resistance under hostile conditions thus becoming a threat globally. Antibiotic resistance is posing a substantial public health threat, hence there is an unmet and urgent clinical need for immune-based non-antibiotic methods to treat brucellosis. Hence, we aimed to explore the whole proteome of Brucella suis to predict antigenic proteins as a vaccine target and designed a novel chimeric vaccine (multi-epitope vaccine) through subtractive genomics-based reverse vaccinology approaches. The applied subsequent hierarchical shortlisting resulted in the identification of Multidrug efflux Resistance-nodulation-division (RND) transporter outer membrane subunit (gene BepC) that may act as a potential vaccine target. T-cell and B-cell epitopes have been predicted from target proteins using a number of immunoinformatic methods. Six MHC I, ten MHC II, and four B-cell epitopes were used to create a 324-amino-acid MEV construct, which was coupled with appropriate linkers and adjuvant. To boost the immunological response to the vaccine, the vaccine was combined with the TLR4 agonist HBHA protein. The MEV structure predicted was found to be highly antigenic, non-toxic, non-allergenic, flexible, stable, and soluble. To confirm the interactions with the receptors, a molecular docking simulation of the MEV was done using the human TLR4 (toll-like receptor 4) and HLAs. The stability and binding of the MEV-docked complexes with TLR4 were assessed using molecular dynamics (MD) simulation. Finally, MEV was reverse translated, its cDNA structure was evaluated, and then, in silico cloning into an E. coli expression host was conducted to promote maximum vaccine protein production with appropriate post-translational modifications. These comprehensive computer calculations backed up the efficacy of the suggested MEV in protecting against B. suis infections. However, more experimental validations are needed to adequately assess the vaccine candidate’s potential.
Highlights
• Subtractive genomic analysis and reverse vaccinology for the prioritization of novel vaccine target
• Examination of chimeric vaccine in terms of allergenicity, antigenicity, MHC I, II binding efficacy, and structural-based studies
• Molecular docking simulation method to rank based vaccine candidate and understand their binding modes
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brucellosis is an endemic disease, also known as Malta fever, Mediterranean fever, undulant fever, and Bang’s disease caused by Brucella genus belong to a family of Brucellaceae, class Alphaproteobacteria infecting both animals and humans [1]. Species of this genus are Gram-negative intracellular facultative pathogens. Based on specific phenotypes with the host and environmental adaptation, till now more than ten known species of Brucella are identified [2]. Brucellosis is characterized as acute fever illness [3], associated with various symptoms in human such as liver and spleen disorders, reproductive abnormalities, neurological problems, heart-related problems, and also have been classified as a potential bioterrorism agent [4, 5]. Brucellosis remains endemic in various emerging countries in Asia, Africa, Middle East, and South America, where screening of livestock and vaccination fails to control and exterminate the disease [6].
The World Health Organization (WHO), Food and Agriculture Organization (FAO), and World Organization for Animal Health (OIE) consider brucellosis as one of the most contagious zoonotic disease worldwide [7], yet human brucellosis remains the commonest zoonotic disease around the world [7]. Brucellosis is transmitted to humans via several routes including, eating raw dairy products from infected animals, aerosol inhalation of an infected animal in clinical lab and slaughterhouse, handling, and exposure to tissue and body fluid of infected animals without Personal Protective Equipment (PPE) [8]. According to Hull [9], there are 500,000 cases of human brucellosis reported per year around the globe due to their ability to survive and multiply within the host phagocytotic and non-phagocytotic cell. Surprisingly, Brucella did not show classical virulence mechanisms such as producing cytolysin, plasmids, fimbria, exotoxin, exoenzymes, and drug resistant forms. However, Brucella is having major virulence factors such as lipopolysaccharide (LPS) [10], β-cyclic glucan [11], outer membrane proteins (Omps) [12], MucR [13], T4SS secretion system, and BvrR/BvrS system which permit the Brucella to interact with the host cell [14]. Similarly, T4SS-based VirB proteins complex their 5 effectors are an essential part of Brucella pathogenesis that regulate the host cell inflammation response and vesicle trafficking [15]. Due to the limited knowledge regarding the genomics of Brucella suis, a new species-specific therapeutic compound and vaccine candidates are difficult to design experimentally [16]. The immune system prophylactic and designing of new therapeutic approaches are of significant interest to combat the antimicrobial resistance. The peptide-based chimeric vaccine, expressed by the pathogenic strain of Brucella, would be an appropriate alternative to its resistance.
Nevertheless, evaluation of thousands of macro-molecules and their subsequent in vivo assays in the wet lab need a lot of time and cost for vaccine design. On the other hand, development in computational biology and various other bioinformatics fields have made wonderful progress leading to a great reduction in consumption of time and associated expenses [17,18,19]. Bioinformatics analysis typically employs alternative approaches for finding novel drug targets, identifying vaccine or drug candidates, elucidating the host–pathogen interactions, designing structure-based drugs, allowing genome-based comparative study, and so on. It is thereby reducing the conventional laboratory-based experimental practice [20,21,22]. The subtractive genomic and reverse vaccinology are one of the most used computational approaches to evaluate the suitability of the vaccine target among the previously known drug target based on selectivity and specificity. Numerous studies had widely reported the use of subtractive genomics and reverse vaccinology approaches against various pathogenic strains for the identification of novel species-specific therapeutic targets [22,23,24,25]. Hence, in the current study, we used the subtractive genomic analysis and reverse vaccinology approach against the whole proteome of the Brucella suis strain to shortlist the vital proteins as vaccine targets. The results of this study suggest that our resultant proteins may be considered the best vaccine targets along with identified multi-epitopes chimeric vaccine that can be used for establishing a universal vaccine, which may provide a basic pipeline against Brucellosis.
Material and methods
The current study utilized a subtractive genomic and reverse vaccinology approach for the identification of Brucella suis specific vaccine targets [26].
Data retrieval
The complete proteome of Brucella suis was retrieved from the National Centre for Biotechnology Information (NCBI) database [27] along with the complete proteome of Human from UNIPROT database (Table 1) [28]. The Database of Essential Gene (DEG) was used to identify the essentiality of proteins, whereas KEGG sever was used to retrieve human and Brucella metabolic pathways.
Prioritization of pathogen-specific metabolic pathways
The KEGG [29] and Automatic Annotation Server (KAAS) metabolic pathway database were used for the analysis of human-host and Brucella suis metabolic pathways. We retrieved the metabolic pathway ID with related information from the KEGG database. Those pathways which were present in both considered a common metabolic pathway, while the remaining considered unique metabolic pathways. We retrieved the FASTA sequence of proteins involved in unique metabolic pathways of the Brucella suis from the NCBI database.
Non-homologous protein identification
Unique metabolic pathways proteins were subjected to evaluate through a BLASTp against the whole proteome of a human using the cut-off value of 0.0001 (E-value 10–3). Those proteins which had a high sequence similarity (> 80%) with human proteome were excluded and the remaining proteins having no similarities were retrieved and further analyzed in the next step of subtractive genomic analysis.
Prioritization of essential proteins among the whole proteome of Brucella suis
In order to prioritize the essential proteins of the Brucella suis the proteins were analyzed via BLASTp with a threshold value of 10–5, against the Database of Essential Gene (DEG) [30] consisted of proteins responsible for the survival of the organisms. Those proteins which had sequence similarity with essential protein of DEG were further analyzed and the non-essential proteins were excluded.
Determination of virulence factor (proteins)
The virulence of proteins helps bacteria to destroy the host-immune system with the help of colonization and invading the host immune cell and as a result, the disease caused. For the determination of virulence of proteins, VFDB (virulence factor of pathogenic bacteria) online database [31] was used. The shortlisted proteins of Brucella suis were subjected to BLAST against the VFDB.
Identification of resistance proteins
The Antibiotic Resistance Gene-ANNOTation V6 (ARG-ANNOT V6) tool was used for the prediction of novel resistance protein sequences from whole-genome and proteome of a pathogen. All the resistant proteins data were collected and analyzed from different experimental published work, different online sources, and protein sequences were retrieved from NCBI database. The shortlisted proteins FASTA sequence were then subjected to BLAST against the resistance proteins of the ARG-ANNOT V6 database with a threshold of 10–5 [32].
Subcellular localization of shortlisted proteins
All the shortlisted proteins were then subjected to PSORTb version 3.0.2 [33] and Cello v.2.5 [34] online tool in order to identify subcellular localization. The main principle of subcellular localization (SCI BLAST) is to BLAST all the shortlisted proteins against the PSORTb and Cello v.2.5 online servers. The subcellular localization consists of cytoplasm, cytoplasmic membrane, periplasmic membrane, and extracellular space, and some are unknown.
Non-homolog gut protein identification
Nonhomologous essential proteins selected as vaccine candidates and novel drug targets were further subjected to standalone BLASTp with an E value cutoff score of 0.001 against the data set present in the Human Microbiome Project server (https://www.hmpdacc.org/hmp/) “28,331” (BioProject) retrieved as gastrointestinal tract from HMP [35] i.e., (https://hmpdacc.org/hmp/catalog/grid.php?dataset=genomic&hmp_isolation_body_site=gastrointestinal_tract) having 26,295 proteins (accessed on 1st Sep, 2022). The HMP sampled microbes from a healthy adult population of 239 people, taking samples from 18 different sites around the body (the mouth, skin, nose, intestines, and genitourinary tract). This yielded ~ 823 unique gut microbial species compositions [36]. Novel therapeutic targets and vaccine candidates were found for proteins with a no similarity threshold. Based on these findings, precautions may be taken to ensure that human microflora proteins are not inadvertently inhibited or blocked.
Prediction of antigenic protein
Outer membrane protein shortlisted from the above subtractive genomic analysis was selected that contain the potency for vaccine development. A bioinformatics approach was applied to that protein to identify the epitopes to boost the immune response of the host. For the prediction of antigenic outer membrane protein VaxiJen webserver [37] was used with a default parameter of 0.5 considered potent antigenicity.
Prediction of MHC class I T-cell epitope
The MHC I epitopes for the selected protein was predicted by NetCTL server [38]. The predicted epitopes were chosen based on a high score. The default parameter of 0.75 was used for the prediction of potent T cell MHC I epitopes.
Prediction of MHC I epitope immunogenicity
A bioinformatics tool IEDB server [39] was used for MHC I antigenicity prediction. The MHC I epitopes should have the strength to evoke a host immune response. A default parameter was used to predict the immunogenicity of MHC I epitopes. Those epitopes having positive antigenic values were selected for further analysis.
Antigenicity, conservancy, and toxicity analysis
The immunogenic epitopes obtained from the IEDB server were analyzed for the antigenic capability using Vaxijen version 2.0 server with a threshold value of 0.5. The IEDB conservancy analysis [40] tool was used for the assessment of the conserved sequence among all the genotypes of MHC I epitopes. The conserved sequence identity parameter was set as a default. This analysis showed the conserved epitopes within the given protein sequence [41]. The assessment of toxicity level was predicted by an online tool called ToxinPred. The parameter was set on default. The toxicity level confirms that the specific host immune response will only target the bacteria rather than host cell itself [42].
MHC II epitope prediction
The IEDB-AR server was used for the identification of MHC class 2 epitopes. It applies the consensus-based prediction approach of both the average relative binding matrix method and stabilization matrix alignment method [43].
MHC I- and II-restricted allele cluster analysis
To confirm the predicted T-cell epitopes, the MHCcluster v2.0 [44] was used to find the cluster of MHC-restricted alleles with their appropriate peptides. This server crosschecks the MHC-restricted allele analysis from the IEDB analysis resources. It results in the static heat map and phylogenetic tree analyzing the functional relation between peptides and HLAs.
Prediction of B-cell epitopes
The B-cell epitope prediction was performed by online bioinformatics server BCPred [45] and FBCpred server [45]. The BCPred works on five different kernel methods whereas, FBCpred is based on consequent kernel methods. The cut-off score of 0.8 was used for B-cell epitopes identification via BCPred [46]. The IEDB B-cell epitope prediction server was used for biochemical properties analysis such as hydrophobicity, surface accessibility, hydrophilicity, amino acid composition, and secondary structure to predict the linear B-cell epitopes.
Construction of model vaccine
Different combinations of the shortlisted T-cell and B-cell epitopes were conjugated sequentially to model a vaccine construct with low toxicity, allergenicity, and high immunogenicity. During the vaccine construction, four different epitopes sequence were added in different combinations. Different amino acid linkers such as GGGS were used between these sequences. To enhance the antigenicity of the vaccine, PADRE (Pan HLA-DR reactive epitope) along with four different adjuvants such as HBHA, HBHA conserved, ribosomal, and beta-defensin were used in each vaccine construct, respectively. The PADRE sequence induced CD4 + T-cells that improve efficacy and potency of peptide vaccine [47]. The adjuvant HBHA and ribosomal adjuvants sequence are agonists of the toll-like receptor 4 (TLR4) while beta-defensin adjuvant is an agonist of the TLR1, TLR2, and TLR4. The sequence-fused vaccine construct was used for further analysis.
Evaluation of allergenicity, antigenicity, and solubility for vaccine construct
The allergenicity was determined by online tool AlgPred program (Saha & Raghava, 2006). The AlgPred tool uses all these parameters (IgE epitope + MAST + ARPs BLAST + SVM) combined to predict the allergenicity of vaccine construct with a threshold value of − 0.40 prediction score. The ANTIGENpro program [48] was used for the determination of vaccine antigenicity while SOLpro was used for the solubility of a vaccine.
Physicochemical properties of vaccine construct
The physicochemical properties of vaccine sequence were observed by the Expasy ProtParam online server [49]. This tool is used for the identification of different physicochemical properties such as molecular weight, number of amino acids, PI values, hydropathicity GRAVY values, aliphatic index, instability index, and estimated half-life of the protein of a generated protein model. This software can analyze various physicochemical properties based on pKa values of different amino acids sequence of a vaccine. The instability index of protein forecasts whether our vaccine is stable or unstable. The aliphatic index of a protein referred to the volume occupied by the aliphatic side chains of amino acid.
Secondary structure modeling
The modeling of vaccine construct was performed by the online tool Swiss modeler [50]. The homology models are constructed using the automated SWISS-MODEL homology modeling server pipeline, and experimental resolve crystal structures of proteins are mapped between the PDB database and UniProtKB using SIFTS [51]. The PSIPRED and PROCHECK were used for the model structure evaluation. The Psi-BLAST selects the relating sequence which had similarity > 80.6% with reported proteins. The best model for each protein was selected and subjected to further analysis.
Molecular docking and molecular dynamic simulation
The molecular docking of V1 with 6 different human leukocyte antigen (HLA) alleles was performed via online tool PatchDock [52] to show the interaction of V1 with HLA alleles. Six different HLA alleles were downloaded from the PDB database with its PDB ID 2FSE (HLA-DRB1*01:01), 3C5J (HLA-DR B3*02:02), 1H15 (HLA-DR B5*01:01), 2Q6W (HLA-DR B3*01:01), 2SEB (HLA-DRB1*04:01), and 1A6A (HLA-DR B1*03:01). The FireDock (Fast Interaction Refinement in Molecular Docking) was used for further validation and more refinement of interactions [52]. Similarly, docking of vaccine (V1) with TLR4 was performed by the GRAMMX [53]. Finally, molecular dynamic simulation of vaccine and TLR4 complex was performed using GROMACS [54]. The final vaccine solvation was executed with an SPC water model in a cubic box with energy minimization using the steepest algorithm. Here, the system was firstly equilibrated using NVT ensemble followed by NPT ensemble. Finally, the vaccine’s molecular dynamic simulation was performed for 10 ns. Furthermore, molecular dynamic simulation of docked complex (vaccine with TLR4) was performed via iMODs server [55]. The iMODs define and calculate the flexibility of protein complex and can be accessed freely. The server asses the direction and extent of the immanent motions of the complex in terms of B-factors, deformability, covariance, and eigenvalue.
Optimization and in silico cloning of final vaccine model
The amino acid sequence of vaccine was reversed translated into DNA sequence to optimized and enhance the expression of chimeric vaccine proteins in the E. coli system through JCAT (Java Codon Adaptation Tool) [56]. The JCAT tool resulted in the prediction of GC content and codon Adaption Index score (CAI) for DNA sequences while avoiding the cleavage site for restriction enzymes Rho-independent termination of transcription and prokaryotic ribosome binding sites. Snapgene tool was used for the insertion of an optimized amino acid sequence of vaccine construct in E. coli pET-28a( +) expression system.
Results
Unique metabolic pathway analysis
Complete metabolic pathways of both Brucella suis (109 pathways) and human host (330 pathways) were downloaded from the KEGG (Kyoto encyclopedia of gene and genome) server. We compared both the human and Brucella suis metabolic pathways manually to find both the common and unique metabolic pathways. The results showed that 71 pathways were common (Table S1) both in humans and Brucella suis and the 38 pathways (Table 2) were unique to Brucella suis. Among these 38 pathways, two-component system, quorum sensing, ribosome, flagellar assembly, and bacterial secretion pathways was observed as highly unique pathways to brucella with 64, 114, 55, 35, and 27 unique proteins. Two-component system (TCS) and quorum sensing regulate the expression of virulence genes, enable the pathogen to detect changes in its external environment and adapt its gene expression appropriately [57]. For example, the quorum-sensing (QS) regulator VjbR, which triggers virB expression, is encoded by the gene encoding the two-component regulator BvrRS, which enables the brucellae to sense the acidic pH and food deprivation they face in the eBCV [57]. In Brucella, a non-motile bacterium, the flagellum, is involved in virulence, infectivity, cell proliferation, and biofilm formation. Brucella’s flagellar proteins are expressed, providing evidence for an evolutionary scenario in which a free-living bacterium acquired flagellar genes from environmental microorganisms, conferring on it the ability to reach other hosts (mammals), and, under selective pressure from the environment, can express these genes, helping it evade the immune response [58]. Likewise, the bacterial secretion pathways and ribosomal pathways help brucella to evade certain non-favorable environment by the secretion of various toxins and proteins. Additionally, Methane metabolism, Lysine biosynthesis, Lipopolysaccharide biosynthesis, O-Antigen nucleotide sugar biosynthesis, Peptidoglycan biosynthesis, Benzoate degradation, Beta-Lactam resistance, and Cationic antimicrobial peptide (CAMP) resistance pathways were consisting of 10–20 unique proteins. These metabolic pathways enable brucella to modulate the cell wall composition to evade the antibiotic environment. Moreover, other pathways such as Vancomycin resistance, d-Alanine metabolism, Carotenoid biosynthesis, Limonene and pinene degradation, Geraniol degradation, Polyketide sugar unit biosynthesis, Carbapenem, Monobactam, Streptomycin, and Novobiocin biosynthesis having less than 10 unique proteins respectively. Total unique metabolic pathways consisted of 503 proteins (Fig. 1).
Prioritization of non-homologous proteins
We subjected 503 unique metabolic pathways of Brucella suis to run a BLASTp with a cutoff value 10–3 against the whole proteome of humans to identify only non-homologous proteins for novel drug targets prioritization. We selected only non-homologous proteins to avoid the undesirable side effects of the drug. The BLASTp results showed 82 proteins were homologous to human host with high similarity with human proteome and were excluded. The remaining 421 non-homologous proteins were analyzed in next step.
Identification of the essential proteins
The Database of Essential Gene (DEG) provides complete information of the essentiality of proteins of the bacteria determined from the experimental analysis. Those proteins having high similarity with proteins of DEG were selected as essential proteins. We ran a BLASTp of non-homologous 421 proteins against the DEG database with a default parameter 60% sequence identity. The results showed that 350 proteins were essential for the survival of Brucella suis.
Predicting the virulence factor (proteins) of B. suis
The VFDB database comes up with complete information of protein virulence. The VFDB explored the importance of virulent proteins in disease progression. The VFDB results revealed that 45 proteins out of 350 (shortlisted proteins after the analysis of metabolic pathways associated proteins) were correlated with virulence of B. suis. However, these 45 proteins can be used as a novel and potent drug target against B. suis strain 1330.
Identification of the resistance proteins
The pathogens resistant to drugs are more hassle to treat the disease, which requires a higher dose that shows a diverse effect in patients. The pathogen acquires with a drug resistance is due to the continuous exposure to drug or the drug used in a higher dose. We subjected the FASTA sequence of shortlisted virulent proteins to ARG-ANNOT V6 online tool. The results showed that 42 out of 45 proteins were correlated with the resistivity of a pathogen. These 42 proteins are mostly involved in the degradation and efflux of numerous drugs. However, these 45 proteins can be used as a potential drug target.
Prediction of subcellular localization
All proteins require a specific location for their optimal function. Transporting proteins to unspecified region may result in server diseases [59]. Based on the subcellular localization of proteins, we design vaccine and drug against the specific localized protein target. The cytoplasmic proteins may act as drug target and outer membrane proteins may act as vaccine target. The results of current study showed ~ 23 cytoplasmic, ~ 14 periplasmic, ~ 6 outer membrane, and ~ 4 inner membrane proteins. Figure 2 represents the graphical sketch of sub cellular localization.
Non-homolog gut protein identification
Different beneficial activities of the human microbiome were reported, and the link between gut flora and humans is not only commensal but symbiotic, mutualistic [60]. The host may experience negative consequences if proteins in this microbiota are blocked or inhibited inadvertently. Therefore, the BLATp was performed against the human gut microbial strains included in the HMP server, and the results indicated that only 2 of 42 proteins exhibited no similarity i.e., sn-glycerol-3-phosphate ABC transporter substrate-binding protein UgpB (WP_004686048.1, Periplasmic Protein), and multidrug efflux RND transporter outer membrane subunit BepC (WP_011068960.1, outermembrane). Because these proteins are not part of shared host–pathogen pathways and have no homology with human “anti-targets,” they are an appropriate target.
Prediction of antigenic protein
The antigenicity of identified outer membrane proteins (n = 6) were predicted to prioritize potential vaccine target against B. suis i.e., d-alanyl-d-alanine carboxypeptidase, Amino acid ABC transporter substrate-binding protein, and multidrug efflux RND transporter outer membrane subunit BepC. We uploaded these protein sequences to VaxiJen web server for the identification of antigenicity of outer membrane protein. The Vaxijen results characterized BepC as the most antigenic protein on the basis of antigenic prediction score which is 0.6511 making protein probable antigenic in nature. It was the only protein fully characterized as outermembrane protein from both the tools (Psortb and cello2) (Table 3). Out of six shortlisted proteins from the subtractive genomic analysis approach, we selected the outer-membrane protein Multidrug efflux Resistance-nodulation-division (RND) transporter outer membrane subunit (BepC gene) for the designing of multi-epitope vaccine. Hence, we may propose this protein for the designing of vaccine.
Prediction of T-cell MHC-I epitopes
The prediction of T-cell MHC-I was achieved by bioinformatics online tool NetCTL server by uploading the FASTA sequence of BepC protein into it. The NetCTL server predicted 448 T-cell epitopes in BepC protein. We selected 83 T-cell epitopes on the basis of high scores than a threshold of 0.2. Epitope’s sequence having a high predicted score representing the higher capability. These 83 predicted epitopes were then subjected to IEDB server to predict the binding affinity of these epitopes to MHC Class I. It resulted in the identification of 35 peptides with binding interaction with MHC1 molecules (Table S2).
MHC I epitope immunogenicity prediction
The epitopes present on the MHC molecules are recognized by CD + 8 to detect the aberrancies such as an infection. Several studies have shown that some peptides are more immunogenic than another peptide because of their amino acid sequence such as peptide having more aromatic amino acids are more immunogenic than other. The strength of the interaction between the peptide-MHC complexes (pMHC) TCRs depends both on the MHC I molecules and the presented peptide. The capability of epitope to stimulate T-cell responses depends on the level of immunogenicity score. The resultant epitopes from the above analysis were further subjected to IEDB server to determine the immunogenicity with a cut-off value of the positive score. The IEDB results showed that out of 35 epitopes 15 epitopes (after the removal of redundant sequences) were most immunogenic. Hence, we selected these immunogenic epitopes for further analysis as shown in Table 4.
Antigenic, conservancy, and toxicity analysis
For the assessment of toxicity, the level of toxicity was predicted by an online tool called as ToxinPred. The results showed that all selected 15 epitopes were not toxic to the host cell. Hence, we selected these epitopes for further analysis. Similarly, IEDB conserved sequence analysis tool was used for the assessment of the conserved sequence among all the genotype of MHC I epitopes. The conserved sequence identity parameters were set as a default. This analysis showed the conserved epitopes within the given protein sequence. The epitopes which showed 50% conserved sequence were selected as a conserved epitope. The result showed that all the 15 epitopes were shown 100% conserved sequence. Non-toxic and most conserved epitopes as a result of the ToxinPred and IEDB tool respectively were further analyzed for their antigenicity through VaxiJen with a cut-off value of 0.5. The results of Vaxijen server showed that 6 epitopes were found depicting the most antigenicity (Table 5). Hence, we selected these 6 epitopes as MHC I epitopes and these are DVKTAEATY, NVAAAETQV, MLFDGFQTR, ALSETLTGA, AMNEQVRAA, and NTASIGVGV.
Prediction of MHC II epitopes
In addition to MHC Class I epitope prediction, the BepC protein was used to predict MHC Class II using IEDB server. The epitopes having binding affinity < 200 nM and percentile ranks < 0.2 were shortlisted and used for further analysis. The results showed that total 11,934 epitopes were generated. We further shortlisted 10 epitopes by using the cut off value of 0.2 i.e., RSTAIAALNAARADV, SRSTAIAALNAARAD, STAIAALNAARADVK, TAIAALNAARADVKT, AIAALNAARADVKTA, ASRSTAIAALNAARA, CKELVAAAVLLSGTV, KELVAAAVLLSGTVL, ACKELVAAAVLLSGT, and KACKELVAAAVLLSG as shown in Table 6.
MHC restriction cluster analysis of shortlisted epitopes
Clusters of MHC-restricted allele and their appropriate peptides were analyzed through MHCcluster v2.0 for the confirmation of the predicted T-cell epitopes on the basis of the IC50 value. Moreover, the interacted alleles were re-evaluated by cluster analysis, and results are shown as a heat map (Fig. 3) and phylogenetic tree (Fig. S1) of MHC-1 and MHC II, respectively. Epitopes clustered are formed based on their interaction with the human leukocyte antigen (HLA). The yellow color shows weaker interactions whereas red color represents strong interactions with proper annotation.
B-cell epitope prediction in BepC
In order to inflict humoral immunity, apart from the MHC-I and MHC-II epitopes (cellular immunity), B-cell epitopes were also identified using BepC protein sequences. For the elimination of pathogen, humoral immunity is also needed along with cellular immunity. The B-cell epitope prediction and classification plays a vital role in vaccine designing, antibody production, and immunodiagnostic tests. Identifying the B-cell epitopes experimentally is an expensive and time-consuming process while computational methods are highly desirable to predict the B-cell epitopes. Hence, the prediction of B-cell epitopes was performed using BCPreds, FBCpred, and ABCpred tools. The results showed that five epitopes were generated via BCPred server, twelve epitopes were generated through the FBCpred tool and twenty-eight epitopes were predicted via ABCpred server (Table S3).
Predicted epitopes comparison for vaccine construct
The predicted B-cell epitopes were manually compared and aligned against MHC I-II epitopes for the construction of the final chimeric vaccine designing. For the final vaccine models, the epitopes having sequences consists of overlapping T- and B-cell epitopes were selected. Finally, we shortlisted four peptides on the basis of their similarities among MHC-I, MHC-II, and B-cell epitopes i.e., GIQLNQMLFDGFQTR, DTIAGTDMGDGNTASIGVGV, TVFKACKELVAAAVL, and AQAEASRSTAIAALNA (Table 7).
Construction of final vaccine model
An adjuvant and PADRE sequence (AKFVAAWTLKAAA) are the most significant component of a multi-epitope vaccine that inflict strong immune response in the host body [61, 62]. The PADRE sequences were chosen for the construction of vaccine in order to examine the adjuvants effects on the antigenicity and allergenicity and to combat the polymorphism complications of HLA-DR molecules caused in the global population. A total of sixteen vaccine constructs were joined with the EAAAK linker and respective adjuvants (HBHA, HBHA conserved, ribosomal, and beta defensin). The MHC Class-I, MHC Class-II, and B-cell epitopes were linked with GGGS, HEYGAEALERAG, and PADRE sequence. A study reported that all the epitopes were joined together with these two linkers do not change the conformation of designed vaccine construct [63] (Table S4).
Prediction of antigenicity, allergenicity, and solubility for vaccine models
The prediction of non-allergic vaccine constructs was performed by AlgPred online server. The constructed vaccine should not be allergenic, because allergic vaccines can induce a cross-reaction in the body. The allergenicity of all the sixteen vaccine constructs was checked and only those vaccines were selected which showed a value > 0.7. Similarly, the antigenicity and solubility of the designed multi-epitope vaccines were also determined by ANTIGENpro and SOLpro, respectively. The solubility of all the vaccine constructs was > 0.8. Finally, we selected only one vaccine to construct on the basis of allergenicity, antigenicity, and solubility for further analysis (Table S5).
Physicochemical analysis of vaccine constructs
The physicochemical properties of vaccine constructs were assessed by ProtParam server which includes a number of amino acids, molecular weight, aliphatic index, PI value, hydropathicity index, and instability index of the shortlisted vaccine constructs. The ProtParam results showed that the molecular weight of molded vaccine ~ 33 kDa, PI was calculated to be 5.03, having stability with 26.98 scores while GRAVY index was calculated to be −0.160 (Table S6).
Construction of 3D structure of V1
We needed the 3-dimensional (3D) structure of vaccine construct to be functional. Hence, we modeled the 3D structure of vaccine via online tool Phyre2. We uploaded the FASTA sequence of vaccine to Phyre2. It compares the FASTA sequence of vaccine with earlier reported 3D structures in PDB database. The Phyre2 results provided us different modeled structures against different template proteins. Finally, we selected the modeled protein against the lipid-binding protein Ce-FAR-7 (2w9y) on the basis of percent identity and confidence score and modeled the vaccine structure (Fig. 4).
Structure validation through Ramachandran plot (PROCHECK)
The constructed 3D structure of the vaccine was validated by online server Procheck. The Procheck results revealed that 93.2% of residues found in the most accepted regions, and 6.8% residues found at additional allowed regions (Fig. 5a). The secondary structure (β-sheets, α-helices, and random coils) of constructed vaccine was analyzed PSIPRED. The results showed the same number of β-sheets, and α-helices in vaccine construct as modeled by Pyre2 tool (Fig. 5b).
Molecular docking and molecular dynamic simulation
A vaccine construct may have potency to build immune response against a different number of epitopes recognized by HLA allele’s proteins. Therefore, molecular docking of V1 with six different alleles such as 2FSE (HLA-DRB1*01:01), 3C5J (HLA-DR B3*02:02), 1H15(HLA-DR B5*01:01), 2Q6W (HLA-DR B3*01:01), 2SEB (HLA-DRB1*04:01), and 1A6A (HLA-DR B1*03:01) was performed through PatchDock and re-defined by FireDock analysis as shown in Table 8. Similarly, docking of TLR4 and design vaccine was performed by GRAMMX tool. The GRAMXX results showed different interactions of vaccine amino acids with TLR4 protein as shown in Fig. 6.
Docked vaccine construct with TLR4/MD. A Docked complex of vaccine (red) and TL4/MD (purple) (B), interaction occurs between the vaccine model and TLR4/MD protein. Interacting residues of vaccine are represented in orange color, while protein-interacting residues highlighted in blue color, C all interactions found between the docked complexes i.e., blue lines represent hydrogen bonding, red color represents salt bridges
Finally, molecular dynamic simulation of V1 performed by GROMACS tool showed the stability of vaccine models at 7 ns (Fig. 7a).
Moreover, NMA stability of complex simulation resulted in the deformation graph illustrating the peaks (Fig. 8a). The eigenvalue detected for the complex was 1.08615e − 04 as shown in (Fig. 8b). The cumulative variance (green colored) and individual variance (red colored) are displayed by variance and B-factor graph visualizing the relation of the docked complex (Fig. 8C). The covariance map represents the motion between pair of residues of complex, where red color indicates the correlated motion, white color represents uncorrelated motion, and anti-correlated motion is represented by blue color (Fig. 8D). The complex’s elastic map shows the relation between the atoms and darker gray regions, indicating stiffer regions (Fig. 8E).
The results of molecular dynamics simulation of vaccine construct and TLR4/MD docked complex. A Deformability, B eigenvalues, C variance (red color indicates individual variances and green color indicates cumulative variances), and D co-variance map (correlated (red), uncorrelated (white), or anti-correlated (blue) motions)
Codon optimization and cloning of chimeric vaccine construct (V1)
The codon optimization of chimeric vaccine was performed by online tool JCAT. The vaccine FASTA sequence was reverse translated into a 950 base DNA sequence. The JCAT calculated the CAI score which was 1 and the GC content was 70.78% indicating the higher level of expression. The pET28a vector was used for its heterologous cloning and expression in E. coli by the Snapgene tool (Fig. 9).
Discussion
Understanding the proteome of a pathogen is important as it facilitates further comprehensive analysis of proteins in various biochemical and pathological pathways that help in the identification of novel drug targets. The development of a novel therapeutic target and vaccination is a significant scientific challenge to combat Brucella suis-related brucellosis [64]. Peptide-based vaccines are now possible to design due to the advancement of sequence-based technologies, computational analysis, and the abundance of genomes and proteomics data for many diseases [61, 62]. The subtractive genomics along with reverse vaccinology can aid in arraying the vast information regarding genomics and proteomics of various pathogens providing acceleration in drug and vaccines designing and pharmacogenomics in the treatments of bacterial infection. The “vaccinomics/Reverse Vaccinology” [65] approach has been proved to be promising approach widely used against Meningococcus B (MenB) [66], antibiotic-resistant Staphylococcus aureus [67], Chlamydia [68], group A Streptococcus [69], and Streptococcus pneumonia [70].
Brucellosis is characterized as acute fever illness associated with various symptoms in human such as liver and spleen disorders, reproductive abnormalities, neurological problems, heart-related problems, and also have been classified as a potential bioterrorism agent. There are 500,000 cases of human brucellosis reported per year around the globe due to their ability to survive and multiply within the host phagocytotic and non-phagocytotic cell. Due to the limited knowledge regarding the genomics of Brucella suis, new species-specific therapeutic compound and vaccine candidates are difficult to design experimentally. Therefore, the development of new vaccines model against Brucella suis is necessary.
In the present study, we have applied reverse vaccinology and subtractive genomics based computational scheme to screen whole proteome of Brucella Suis for the identification of novel drug target and multi-epitope vaccine construction. We selected unique metabolic pathways composed of proteins unique to Brucella suis and excluded the common metabolic pathways present both in brucella and human-host since unique proteins are of interest. Total unique metabolic pathways consisted of 503 proteins followed by the foretelling of non-homologous protein from the complete proteome of Brucella Suis. Similarly, the essential proteins from the complete proteome of Brucella suis were determine which can be used as a potential drug target. The DEG analysis led to the identification of essential protein required for the survival of Brucella suis followed by the determination of virulence protein which are responsible for infection in human. Likewise, resistance protein identification was also performed. Resistance proteins are those which are responsible for the efflux of various antibiotic from the bacterial cell to counteract the drug action [71]. The subcellular localization of shortlisted proteins was also predicted for the identification of cytoplasmic protein for the drug targets and cell membrane protein for the construction of multi-epitope vaccine [72]. We shortlisted one protein namely Multidrug Efflux RND Transporter Outer Membrane Subunit BepC (involve in LPS metabolic pathway) for vaccine construction. Since this metabolic pathway is essential for bacteria but is absent in humans, BepC is being investigated as a possible vaccine candidate [73]. Surprisingly, Brucella did not show classical virulence mechanisms such as producing cytolysins, plasmids, fimbria, exotoxin, exoenzymes, and drug resistant forms. However, Brucella is having major virulence factors such as, lipopolysaccharide (LPS) [10]. This research employed a combination of prediction techniques to identify putative B- and T-cell epitopes that might trigger humoral or cell-mediated responses. Antibodies, also called immunoglobulins, are produced in large part by a kind of cell called a B-lymphocyte. Each of these epitopes takes on a linear or conformational shape [74]. Only in the protein’s basic structure can the linear B-cell fold be found in its entirety. It is protein folding that brings together discontinuous or conformational B-cell epitopes. This is an essential factor while developing vaccines [75, 76]. As a result of their ability to recognize and interact with MHC (Major Histocompatibility Complex) linked to antigen-presenting cells, T cells are classified as CD4 + and CD8 + cells, respectively (APCs). In summary, these APCs have surface antigens that are recognized by T-cell receptors [73]. The predicted peptide in present study were combined with the help of different linkers and adjuvant to construct chimeric based vaccine against Brucella suis. The addition of used adjuvant increased the immunogenicity of the vaccine formulation. Because of its immunomodulatory characteristics, this adjuvant is increasingly being employed in multiepitope vaccines [72]. Eventually, 16 vaccine constructs were designed against Brucella suis which were comprehensively investigated for toxicity profile, allergenicity pattern immunogenicity, and conservancy analysis resultant in only one final vaccine construct. The interaction of modeled vaccine with human leukocyte antigen (HLA) allele to interpret effective immune response was examine using molecular docking simulation studies with GRAMAXX and GROMACS tools. Protein–protein docking is frequently used in reverse vaccinology to identify the most promising vaccine design [72]. However, the results of docking analysis showed the binding affinity of promiscuous epitopes with different HLA alleles. An important TLR in mammals, TLR2/4 can identify lipoproteins from bacteria, viruses, fungi, and parasites [77]. Therefore, the shortlisted vaccine construct was used for the binding affinity and stability estimation of the of the vaccine and TLR4 complex. The JCAT tool was used for codon optimization. The JCAT calculated the CAI score which was 1 and the GC content was 70.78% indicating the higher level of expression. The pET28a vector was used for its heterologous cloning and expression in E. coli by the Snapgene tool. In order to confirm the findings of this research, it is recommended that the vaccine candidate be expressed in bacteria for further investigation. The inquiry would next be guided by pre-clinical studies, including tissue-culture or cell-culture systems, and animal experimentation.
Conclusion
The current study applied the subtractive genomics and reverse vaccinology approach for the prioritization of potent vaccine targets against Brucella suis 1300 strain. It smears multiple essential analyses at different stages i.e., non-homologs, essential, and unique to pathogens proteins, and sub cellular localization. In this study numbers of proteins along with Multidrug efflux RND transporter outer membrane subunit BepC was shortlisted as a novel vaccine target against Brucella suis. Consequently, the shortlisted essential proteins may be further study and used as a therapeutic vaccine candidate for Brucella Suis. Furthermore, immunogenicity prediction, allergenicity identification, and tertiary structure analysis of vaccine proposed it as a potent chimeric vaccine against Brucella suis. The molecular docking simulation and codon optimization were also found as satisfactory hence giving confidence to this study. It was found that the designed MEV in the present study can make stable interactions with human immune receptors and able to stimulate an efficient host immune system response. However, experimental validation with computational approaches is required for further analysis to improve the efficacy of predicted MEV.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Moreno E, Cloeckaert A, Moriyón I. Brucella evolution and taxonomy. Vet Microbiol. 2002;90(1–4):209–27.
Pappas G, et al. The new global map of human brucellosis. Lancet Infect Dis. 2006;6(2):91–9.
Posadas DM, et al. BmaC, a novel autotransporter of Brucella suis, is involved in bacterial adhesion to host cells. Cell Microbiol. 2012;14(6):965–82.
Gyuranecz M, et al. Detection of Brucella canis–induced reproductive diseases in a kennel. J Vet Diagn Invest. 2011;23(1):143–7.
Doganay GD, Doganay M. Brucella as a potential agent of bioterrorism. Recent Pat Antiinfect Drug Discov. 2013;8(1):27–33.
Seleem MN, Boyle SM, Sriranganathan N. Brucellosis: a re-emerging zoonosis. Vet Microbiol. 2010;140(3–4):392–8.
Schelling E, et al. Brucellosis and Q-fever seroprevalences of nomadic pastoralists and their livestock in Chad. Prev Vet Med. 2003;61(4):279–93.
Fiori PL, et al. Brucella abortus infection acquired in microbiology laboratories. J Clin Microbiol. 2000;38(5):2005–6.
Hull NC, Schumaker BA. Comparisons of brucellosis between human and veterinary medicine. Infect Ecol Epidemiol. 2018;8(1):1500846.
Lapaque N, et al. Brucella lipopolysaccharide acts as a virulence factor. Curr Opin Microbiol. 2005;8(1):60–6.
Martirosyan A, et al. Brucella β 1, 2 cyclic glucan is an activator of human and mouse dendritic cells. PLoS Pathog. 2012;8(11): e1002983.
Lim JJ, et al. Protective effects of recombinant Brucella abortus Omp28 against infection with a virulent strain of Brucella abortus 544 in mice. J Vet Sci. 2012;13(3):287.
Mirabella A, et al. Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, Is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J Bacteriol. 2013;195(3):453–65.
Głowacka P, et al. Brucella–virulence factors, pathogenesis and treatment. Pol J Microbiol. 2018;67(2):151.
Ke Y, et al. Type IV secretion system of Brucella spp. and its effectors. Front Cell Infect Microbiol. 2015;5:72.
Henriquez-Camacho C, et al. Proteins of Bartonella bacilliformis: candidates for vaccine development. Int J Pept. 2015;2015:1–5.
Barh D, et al. A novel strategy of epitope design in Neisseria gonorrhoeae. Bioinform Biol Insights. 2010;5(2):77.
Lin X, Li X, Lin X. A review on applications of computational methods in drug screening and design. Molecules. 2020;25(6):1375.
Hasan M, et al. Reverse vaccinology approach to design a novel multi-epitope subunit vaccine against avian influenza A (H7N9) virus. Microb Pathog. 2019;130:19–37.
Zhu H. Big data and artificial intelligence modeling for drug discovery. Annu Rev Pharmacol Toxicol. 2020;60:573–89.
Lakshmi P, Ramyachitra D. Review about bioinformatics, databases, sequence alignment, docking, and drug discovery. In: Statistical Modelling and Machine Learning Principles for Bioinformatics Techniques, Tools, and Applications. Springer; 2020. p. 11–23.
Hasan M, et al. Vaccinomics strategy for developing a unique multi-epitope monovalent vaccine against Marburg marburgvirus. Infect Genet Evol. 2019;70:140–57.
Uddin R, Jamil F. Prioritization of potential drug targets against P. aeruginosa by core proteomic analysis using computational subtractive genomics and Protein-Protein interaction network. Comput Biol Chem. 2018;74:115–22.
Sarangi AN, et al. Subtractive genomics approach for in silico identification and characterization of novel drug targets in Neisseria Meningitides Serogroup B. J Comput Sci Syst Biol. 2009;2(5):255–8.
Mondal SI, et al. Identification of potential drug targets by subtractive genome analysis of Escherichia coli O157: H7: an in silico approach. Adv Appl Bioinform Chem: AABC. 2015;8:49.
Uddin R, et al. Computational identification of potential drug targets against Mycobacterium leprae. Med Chem Res. 2016;25(3):473–81.
Tatusova TA, Karsch-Mizrachi I, Ostell JA. Complete genomes in WWW Entrez: data representation and analysis. Bioinformatics (Oxford, England). 1999;15(7):536–43.
UniProt: the universal protein knowledgebase in 2021. J Nucleic Acids Res. 2021; 49, no. D1 (2021): D480–D489.
Kanehisa M, et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–51.
Luo H, et al. DEG 15, an update of the Database of Essential Genes that includes built-in analysis tools. Nucleic Acids Res. 2021;49(D1):D677–86.
Liu B, et al. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47(D1):D687–92.
Gupta SK, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58(1):212–20.
Yu NY, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–15.
Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13(5):1402–6.
Peterson J, et al. The NIH human microbiome project. Genome Res. 2009;19(12):2317–23.
Consutium. A framework for human microbiome research. Nature. 2012;486(7402):215–21.
Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BioMed Central Bioinformatics. 2007;8(1):1–7.
Larsen MV, et al. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BioMed Central Bioinformatics. 2007;8(1):1–12.
Calis JJ, et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol. 2013;9(10): e1003266.
Bui H-H, et al. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BioMed Central Bioinformatics. 2007;8(1):1–6.
Dash R, et al. In silico-based vaccine design against Ebola virus glycoprotein. Adv Appl Bioinform Chem: AABC. 2017;10:11.
Karthik L, et al. Protease inhibitors from marine actinobacteria as a potential source for antimalarial compound. PLoS ONE. 2014;9(3): e90972.
Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2016;54(1):5.6. 1–5.6. 37.
Thomsen M, et al. MHCcluster, a method for functional clustering of MHC molecules. Immunogenetics. 2013;65(9):655–65.
El-Manzalawy Y, Dobbs D, Honavar V. Predicting flexible length linear B-cell epitopes. In: Computational Systems Bioinformatics: (Volume 7). World Scientific; 2008. p. 121–32.
Barh D, et al. In silico subtractive genomics for target identification in human bacterial pathogens. Drug Dev Res. 2011;72(2):162–77.
Ghaffari-Nazari H, et al. Improving multi-epitope long peptide vaccine potency by using a strategy that enhances CD4+ T help in BALB/c mice. PLoS ONE. 2015;10(11): e0142563.
Magnan CN, et al. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26(23):2936–43.
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook; 2005, pp 571–607.
Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–303.
Bienert S, et al. The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Res. 2017;45(D1):D313–9.
Mashiach E, et al. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36(suppl_2): W229–W232.
Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 2006;34(suppl_2): W310–W314.
Pronk S, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–54.
López-Blanco JR, et al. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(W1):W271–6.
Grote A, et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33(suppl_2):W526–W531.
Roop RM, et al. Uncovering the hidden credentials of Brucella virulence. Microbiol Mol Biol Rev. 2021;85(1):e00021-e119.
Coloma-Rivero RF, et al. Brucella and its hidden flagellar system. Microorganisms. 2021;10(1):83.
Shurety W, et al. Localization and post-Golgi trafficking of tumor necrosis factor-alpha in macrophages. J Interferon Cytokine Res. 2000;20(4):427–38.
Khan MT, Mahmud A, Hasan M, Azim KF, Begum MK, Rolin MH, Mondal SI. Proteome exploration of legionella pneumophila to identify novel therapeutics: a hierarchical subtractive genomics and reverse vaccinology approach. Microbiol Spectr. 2022;10(4):e00373-22.
Jalal K, et al. Identification of vaccine and drug targets in Shigella dysenteriae sd197 using reverse vaccinology approach. Sci Rep. 2022;12(1):1–19.
Khan K, Jalal K, Uddin R. An integrated in silico based subtractive genomics and reverse vaccinology approach for the identification of novel vaccine candidate and chimeric vaccine against XDR Salmonella typhi H58. Genomics. 2022;114(2): 110301.
Solanki V, Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci Rep. 2018;8(1):1–19.
Scoffone VC, et al. Vaccines to overcome antibiotic resistance: the challenge of Burkholderia cenocepacia. Trends Microbiol. 2020;28(4):315–26.
Donati C, Rappuoli R. Reverse vaccinology in the 21st century: improvements over the original design. Ann N Y Acad Sci. 2013;1285(1):115–32.
Mora M, et al. Reverse vaccinology. Drug Discovery Today. 2003;8(10):459–64.
Sharma S. Identificationof vaccine candidates against Staphylococcus Aureus: an in silico reverse vaccinology approach (Doctoral dissertation). 2019.
Adu-Bobie J, et al. Two years into reverse vaccinology. Vaccine. 2003;21(7–8):605–10.
Seib KL, Zhao X, Rappuoli R. Developing vaccines in the era of genomics: a decade of reverse vaccinology. Clin Microbiol Infect. 2012;18:109–16.
Talukdar S, et al. Identification of potential vaccine candidates against Streptococcus pneumoniae by reverse vaccinology approach. Appl Biochem Biotechnol. 2014;172(6):3026–41.
Basharat Z, et al. Differential analysis of Orientia tsutsugamushi genomes for therapeutic target identification and possible intervention through natural product inhibitor screening. Comput Biol Med. 2022;141: 105165.
Alotaibi G, et al. Pan genome based reverse vaccinology approach to explore Enterococcus faecium (VRE) strains for identification of novel multi-epitopes vaccine candidate. Immunobiology. 2022;227(3): 152221.
Jalal K, et al. Pan-genome reverse vaccinology approach for the design of multi-epitope vaccine construct against Escherichia albertii. Int J Mol Sci. 2021;22(23):12814.
Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and methods for T-and B-cell epitope prediction. J Immunol Res. 2017;2017:1–14.
Jespersen MC, et al. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45(W1):W24–W29.
Moodley A, et al. Reverse vaccinology approach to design a multi-epitope vaccine construct based on the Mycobacterium tuberculosis biomarker PE_PGRS17. Immunol Res. 2022:1–17.
Ardestani H, et al. In silico and in vivo approaches to recombinant multi-epitope immunogen of GroEL provides efficient cross protection against S. Typhimurium, S. flexneri, and S. dysenteriae. Mol Immunol. 2022;144:96–105.
Acknowledgements
The authors would like to acknowledge the Higher Education Commission of Pakistan for providing financial support under National Research Program for Universities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants and animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jalal, K., Khan, K. & Uddin, R. Immunoinformatic-guided designing of multi-epitope vaccine construct against Brucella Suis 1300. Immunol Res 71, 247–266 (2023). https://doi.org/10.1007/s12026-022-09346-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-022-09346-0