Abstract
Purpose
Assessment of pituitary adenoma (PA) volume and extent of resection (EOR) through manual segmentation is time-consuming and likely suffers from poor interrater agreement, especially postoperatively. Automated tumor segmentation and volumetry by use of deep learning techniques may provide more objective and quick volumetry.
Methods
We developed an automated volumetry pipeline for pituitary adenoma. Preoperative and three-month postoperative T1-weighted, contrast-enhanced magnetic resonance imaging (MRI) with manual segmentations were used for model training. After adequate preprocessing, an ensemble of convolutional neural networks (CNNs) was trained and validated for preoperative and postoperative automated segmentation of tumor tissue. Generalization was evaluated on a separate holdout set.
Results
In total, 193 image sets were used for training and 20 were held out for validation. At validation using the holdout set, our models (preoperative / postoperative) demonstrated a median Dice score of 0.71 (0.27) / 0 (0), a mean Jaccard score of 0.53 ± 0.21/0.030 ± 0.085 and a mean 95th percentile Hausdorff distance of 3.89 ± 1.96./12.199 ± 6.684. Pearson’s correlation coefficient for volume correlation was 0.85 / 0.22 and −0.14 for extent of resection. Gross total resection was detected with a sensitivity of 66.67% and specificity of 36.36%.
Conclusions
Our volumetry pipeline demonstrated its ability to accurately segment pituitary adenomas. This is highly valuable for lesion detection and evaluation of progression of pituitary incidentalomas. Postoperatively, however, objective and precise detection of residual tumor remains less successful. Larger datasets, more diverse data, and more elaborate modeling could potentially improve performance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pituitary adenomas (PA) are a frequent type of intracranial tumor [1]. Endonasal transsphenoidal surgery has established itself as the best option for its treatment in most cases [2]. Its outcome varies greatly with different factors like tumor morphology and the surgeon caseload [3,4,5]. Treatment is indicated in case of functioning PA other than prolactinomas, in case of symptomatic PA or in case of relevant volumetric progression [6]. If surgery is performed, assessment of residual tumor is relevant in order to determine the extent of resection (EOR) [7], though manual segmentation of tumor volumes is likely highly dependent on the rater, especially postoperatively [8,9,10]. Automated analysis of pre- and postoperative imaging could consequently have the potential to provide more objective and precise volumetry.
Semantic image segmentation is a classic machine learning application [11, 12], not only due to the fact that manual segmentation requires considerable amounts of expert time [13, 14]. Convolutional neural networks (CNNs)—and specifically U-Nets—have recently been applied successfully for biomedical image segmentation due to their throughput speed and overall good performance in this task [15].
To the best of the authors’ knowledge, no automated approaches to segment PA pre- and postoperatively for volumetry and resection assessment exist. We hypothesize that a CNN can generate segmentations of PA faster and more objective while maintaining quality of segmentation.
Methods
Data and preprocessing
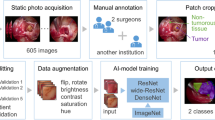
Patients undergoing transsphenoidal surgery for PA at University Hospital Zurich in the period of October 2012 to May 2021 with available preoperative and 3-month postoperative 3-Tesla magnetic resonance imaging (MRI) were included. After identifying the closest T1-weighted contrast-enhanced MRI scan prior to surgery and a three month follow up those were assigned a Study ID and exported. In order to account for different manufacturers and acquisition protocols at referring hospitals, the images were converted to NIfTI format [16], reshaped to 256 × 256 × 256 voxels, voxel size normalized to 1.0 × 1.0 × 1.0, and images were reoriented using a right-anterior-superior affine matrix. Tumor tissue was subsequently manually labeled for training and residual volume assessment. After creating a holdout set of 20 patients for assessment of model generalization, the remaining 386 studies (two per patient) had its pixel intensities normalized for each study individually, and were then sliced in the coronal plane [17].
Model Development
We used a 2D-U-Net as model architecture, with a binary cross entropy loss function, Adam optimizer, and a sigmoid activation function [15]. It was built using the following platforms: Python 3.9.0 [18], Keras 2.5.0 [19], SimpleITK [20,21,22] and nibabel [23]. Training was carried out on a Nvidia RTX 3090 graphical processing unit. Separate preoperative and postoperative models were trained in five-fold cross validation.
The five resulting models were subsequently used to create ensemble segmentations by averaging their respective predictions. To binarize the predicted probabilities ranging from zero to one, a threshold of 0.6 for preoperative and 0.44 for postoperative scans was used as illustrated in Fig. 1. For the postoperative predictions, the following automated postprocessing steps were implemented: Coherent volumes smaller than fifty pixels were removed, holes within the segmentation were filled and a dilate function was used to smoothen the corners which is closer to natural tumor growth.
For the postoperative models, we additionally implemented transfer learning by initializing the postoperative models with the parameters of the fully trained preoperative models, and image augmentation was performed with a sampling ratio of 1/255 and rotation of between 0 and 90 degrees as well as zoom of 0% to 30%.
Evaluation
Manual and automated segmentations were compared using the Dice score, Jaccard score and the 95th percentile of the Hausdorff distance [24,25,26,27]. Dice and Jaccard evaluate similarity and overlap and range from zero—indicating no overlap—to one for perfect congruence. The Hausdorff distance analyzes the distances between two sets of points made up from the edges of two segmentations. Smaller values thus represent better performance. We opted for the 95th percentile instead of maximal Hausdorff distance to decrease the importance of outliers. Volumes were calculated in mm3 from the segmented masks, and their correlation with the manually segmented volumes was assessed using Pearson’s product-moment correlation. Automated and manual EOR were similarly correlated. Finally, we assessed the model’s performance in detecting gross total resection (GTR, defined as an EOR of 100%) using a confusion matrix.
Results
Cohort
In total, 213 patients were included retrospectively, of which 193 were applied for development of the model. Validation was performed in 20 held-out patients. Summary demographics and radiological information are displayed in Table 1.
Preoperative segmentation performance
Segmentation performance is summarized in Table 2. In terms or preoperative tumor segmentation, our ensemble model achieved a mean Dice score of 0.62 ± 0.22, with automatically rated volumes correlating well with manually segmented volumes (r = 0.85). Figure 2 shows metric performance and tumor volume with a linear regression. Exact Jonckheere-Terpstra-Test for a trend did not reach significance (JT, p value for Dice score: 110, 0.1757; JT, p value for Jaccard score: 114, 0.1166) [28].
Postoperative segmentation performance
For postoperative segmentations, a mean Dice score of 0.046 ± 0.125 was observed. The correlation of manually segmented tumors and predicted tumor masks correlated less satisfactory than in preoperative scans (r = 0.22). Introduction of transfer learning techniques and image augmentation did not improve performance (Table 3).
Resection assessment performance
Table 4 summarizes performance in terms of resection assessment. Our model’s predictions generated EOR values that only poorly correlated with manual segmentations (r = −0.14). Automatically detected EOR differed from the ground truth manual segmentation by 18.65% ± 31.10% on average. GTR was detected with an accuracy of 50.00%, sensitivity of 66.67% and specificity of 36.36%.
Discussion
We have developed and validated an automated PA segmentation pipeline based on deep learning. We demonstrate that our approach performs favorably when it comes to segmentation and volumetric assessment of preoperative images. Generating objective and precise postoperative segmentations of residual tumor remains a challenge, even with the application of advanced machine learning techniques.
As neuroimaging has become much more frequent, also detection of incidentalomas is prone to increase since especially nonfunctioning incidentaloma of the pituitary are highly prevalent (1.4–27% in autopsy and 3.7–37% in imaging) [29]. Hence automated segmentation of incidental PA would tackle a frequent issue. Incorporating this into a diagnostic software to detect suspicious lesions of the pituitary gland would be valuable for clinical routine. To address this, we developed a fully automated graphical user interface that tackles this issue (https://micnlab.com/download-the-zurich-neurosurgical-toolkit/).
Furthermore, automated, objective volumetric measurement for assessing progression of incidentalomas, especially microadenomas, yields a clinical benefit since volumetric progression is crucial for surgical indication [30]. Last, prognostic scores like the Zurich pituitary or Knosp score could be automatically implemented, ultimately giving the clinician further information and saving his time. These three options combined have the potential to standardize and speed up the clinical workflow of PA.
When indicated, the transsphenoidal approach is very effective in most cases of PA with comparatively low surgical morbidity and mortality [3]. When evaluating surgical oncology results—not only in individual patients, but also when comparing cohorts, surgeons, and departments and for research purposes—volumetric assessment of residual adenoma volume is of paramount importance. However, segmenting tumors on each slice accurately is time-consuming and often not possible in daily clinical practice. Furthermore, as in other tumor segmentation tasks such as gliomas, the interrater agreement likely is low, especially for postoperative residual tumor tissue [10, 31]. In this light, it must be considered that even morphological grading at the preoperative stage using the Hardy and Knosp classification already suffers from relatively low interrater agreement [8, 9].
Volumetric rating of post-resection sellar scans thus presents a particular challenge when it comes to objectivity. Automated semantic segmentation – the machine learning task that deals with detecting and outlining structures on images—could prove a viable option to improve the speed and objectivity with which volumes are assessed pre- and postoperatively.
To some extent, the poor performance on postoperative images is to be expected: In the end, supervised learning techniques can only ever be as good as the “ground truth” data they are trained on, and with disagreements in labeling of small or debatable residual tumor, this has certainly been the case in this study. Even in the much more intensely studied task of glioma and glioblastoma segmentation, which has been fueled by the yearly international BraTS challenges [32], performance overall appears mediocre and demonstrates that – especially for low grade glioma—it appears to be difficult to generate any improvements in segmentation compared to human raters, apart from the increased speed and objective nature with which automated segmentations can be produced. Even in BraTS 2014/2015, where postoperative images were also segmented, the ground truth labels for postoperative images eventually had to be generated by learning algorithms.
Our ensemble method is—to the best of the authors’ knowledge—the only attempt at automatically segmenting post-transsphenoidal surgery scans. The deep learning approach was able to accurately outline the tumors preoperatively, but struggled with small residual tumors. Certainly, this can be explained at least partially by the computationally necessary downsampling of the images, resulting at times in voxels that appear almost equally as large as the residual tumor itself. Also, pituitary adenomas do not appear constantly with the same relative intensity making them hard to identify at times [33].
Limitations
Although we included scans from many institutions and all major scanner manufacturers, our study remains single-center and external validation would be necessary before any kind of clinical application. Furthermore, we applied 2D segmentation—which has demonstrated reliable results previously for other similar applications—although 3D segmentation potentially could further increase performance. A larger dataset of subtotally resected pituitary adenomas would most likely also allow improvements, since most postoperative images showed no tumor for the model to learn to recognize. A more heterogeneous dataset as we did allows for better generalization and reduces the risk of overfitting to a particular manufacturer or hospital protocol. On the other hand, a reduction in performance is to be expected with this approach.
Conclusion
Our volumetry pipeline demonstrated its ability to accurately and automatically segment pituitary adenoma. This is highly valuable for lesion detection and evaluation progression of pituitary incidentalomas. Postoperatively, however, objective and precise detection of residual tumor remains less successful. Larger datasets, more diverse data, and more elaborate modeling could potentially improve performance. Yet, focusing on use cases for preoperative segmentations seems more promising.
Data availability
The data in support of our findings can be obtained upon reasonable request from the corresponding author. The models can be downloaded and applied using our graphical user interface, available at “https://micnlab.com/download-the-zurich-neurosurgical-toolkit/”.
References
K.A. McNeill, Epidemiology of Brain Tumors. Neurologic Clin. 34, 981–998 (2016). https://doi.org/10.1016/j.ncl.2016.06.014
R. Pratheesh, S. Rajaratnam, K. Prabhu et al. The current role of transcranial surgery in the management of pituitary adenomas. Pituitary 16, 419–434 (2013). https://doi.org/10.1007/s11102-012-0439-z
F.G. Barker, A. Klibanski, B. Swearingen, Transsphenoidal surgery for pituitary tumors in the United States, 1996–2000: mortality, morbidity, and the effects of hospital and surgeon volume. J. Clin. Endocrinol. Metab. 88, 4709–4719 (2003). https://doi.org/10.1210/jc.2003-030461
V.E. Staartjes, C. Serra, M. Zoli et al. Multicenter external validation of the Zurich Pituitary Score. Acta Neurochir. 162, 1287–1295 (2020). https://doi.org/10.1007/s00701-020-04286-w
A.S.G. Micko, A. Wöhrer, S. Wolfsberger, E. Knosp, Invasion of the cavernous sinus space in pituitary adenomas: endoscopic verification and its correlation with an MRI-based classification. J. Neurosurg. 122, 803–811 (2015). https://doi.org/10.3171/2014.12.JNS141083
M. Buchfelder, S. Schlaffer, Surgical treatment of pituitary tumours. Best. Pract. Res. Clin. Endocrinol. Metab. 23, 677–692 (2009). https://doi.org/10.1016/j.beem.2009.05.002
C. Serra, J.-K. Burkhardt, G. Esposito et al. Pituitary surgery and volumetric assessment of extent of resection: a paradigm shift in the use of intraoperative magnetic resonance imaging. FOC 40, E17 (2016). https://doi.org/10.3171/2015.12.FOCUS15564
M.A. Mooney, D.A. Hardesty, J.P. Sheehy et al. Interrater and intrarater reliability of the Knosp scale for pituitary adenoma grading. J. Neurosurg. 126, 1714–1719 (2016). https://doi.org/10.3171/2016.3.JNS153044
M.A. Mooney, D.A. Hardesty, J.P. Sheehy et al. Rater reliability of the hardy classification for pituitary adenomas in the magnetic resonance imaging era. J. Neurol. Surg. B Skull Base 78, 413–418 (2017). https://doi.org/10.1055/s-0037-1603649
M. Visser, D.M.J. Müller, R.J.M. van Duijn et al. Inter-rater agreement in glioma segmentations on longitudinal MRI. NeuroImage: Clin. 22, 101727 (2019). https://doi.org/10.1016/j.nicl.2019.101727
Z. Akkus, A. Galimzianova, A. Hoogi et al. Deep learning for brain MRI segmentation: State of the art and future directions. J. Digit Imaging 30, 449–459 (2017). https://doi.org/10.1007/s10278-017-9983-4
Z. Obermeyer, E.J. Emanuel, Predicting the future — big data, machine learning, and clinical medicine. N. Engl. J. Med 375, 1216–1219 (2016). https://doi.org/10.1056/NEJMp1606181
B.F. King, artificial intelligence and radiology: What will the future hold. J. Am. Coll. Radiol. 15, 501–503 (2018). https://doi.org/10.1016/j.jacr.2017.11.017
J. Egger, T. Kapur, C. Nimsky, R. Kikinis, Pituitary Adenoma Volumetry with 3D Slicer. PLoS ONE 7, e51788 (2012). https://doi.org/10.1371/journal.pone.0051788
Ronneberger O., Fischer P., Brox T. (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv:150504597 [cs]
X. Li, P.S. Morgan, J. Ashburner et al. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 264, 47–56 (2016). https://doi.org/10.1016/j.jneumeth.2016.03.001
Zettler N., Mastmeyer A. (2021) Comparison of 2D vs. 3D U-Net Organ Segmentation in abdominal 3D CT images
Van Rossum G., Drake Jr F.L. (1995) Python reference manual. Centrum voor Wiskunde en Informatica Amsterdam
Chollet F. (2015) Keras. https://github.com/fchollet/keras
Beare R., Lowekamp B., Yaniv Z. (2018) Image Segmentation, Registration and Characterization in R with SimpleITK. J Stat Soft 86. https://doi.org/10.18637/jss.v086.i08
Z. Yaniv, B.C. Lowekamp, H.J. Johnson, R. Beare, SimpleITK Image-Analysis Notebooks: a collaborative environment for education and reproducible research. J. Digit Imaging 31, 290–303 (2018). https://doi.org/10.1007/s10278-017-0037-8
Lowekamp B.C., Chen D.T., Ibáñez L., Blezek D. (2013) The Design of SimpleITK. Front Neuroinform 7. https://doi.org/10.3389/fninf.2013.00045
Brett, M., Markiewicz Christopher J., Hanke, M., et al (2020) nipy/nibabel: 3.2.1
A.A. Taha, A. Hanbury, Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 15, 29 (2015). https://doi.org/10.1186/s12880-015-0068-x
L.R. Dice, Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945). https://doi.org/10.2307/1932409
P. Jaccard, The distribution of the flora in the alpine zone.1. N. Phytol. 11, 37–50 (1912). https://doi.org/10.1111/j.1469-8137.1912.tb05611.x
A. Ralescu, Probability and fuzziness. Inf. Sci. 34, 85–92 (1984). https://doi.org/10.1016/0020-0255(84)90018-5
A.R. Jonckheere, A distribution-free k-sample test against ordered alternatives. Biometrika 41, 133 (1954). https://doi.org/10.2307/2333011
Thapar K., Kovacs K., Scheithauer B., Lloyd R.V. (2001) Diagnosis and management of pituitary tumors. Humana Press
F. Galland, M.-C. Vantyghem, L. Cazabat et al. Management of nonfunctioning pituitary incidentaloma. Annales d’Endocrinologie 76, 191–200 (2015). https://doi.org/10.1016/j.ando.2015.04.004
O. Rodríguez, B. Mateos, R. de la Pedraja et al. Postoperative follow-up of pituitary adenomas after trans-sphenoidal resection: MRI and clinical correlation. Neuroradiology 38, 747–754 (1996). https://doi.org/10.1007/s002340050341
B.H. Menze, A. Jakab, S. Bauer et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med Imaging 34, 1993–2024 (2015). https://doi.org/10.1109/TMI.2014.2377694
K. Karimian-Jazi, Hypophysentumoren. Radiologe 59, 982–991 (2019). https://doi.org/10.1007/s00117-019-0570-1
Acknowledgements
The results of this paper have been presented at the EANS 2022 Congress in Belgrade by Raffaele Da Mutten in a seven-minute presentation on the 18th of October 2022. Consequently, the abstract is available under: https://doi.org/10.1016/j.bas.2022.101287.
Funding
Open access funding provided by University of Zurich.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by R.D.M. and O.C.-C. The code was written and the results were analyzed by R.D.M., O.Z. and V.E.S. The first draft of the manuscript was written by R.D.M. and all authors commented on it. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
The ethics committee of the canton of Zurich approved this project under the BASEC number 2021-01147. No identifying images or data are contained in this text. In this retrospective work, no additional consent was obtained. Before the procedure, consent to the surgery was given.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Da Mutten, R., Zanier, O., Ciobanu-Caraus, O. et al. Automated volumetric assessment of pituitary adenoma. Endocrine 83, 171–177 (2024). https://doi.org/10.1007/s12020-023-03529-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12020-023-03529-x





