Abstract
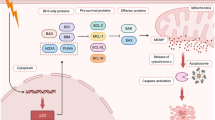
With the absence of the three most common receptor targets, and with high vascularity and higher-grade tumors, triple-negative breast cancer (TNBC) is the most aggressive of all breast cancer subtypes and is in need of additional/alternative/novel treatment strategies. With ~ 15% of the over 2 million new cases each year, there is an unmet need to treat TNBC. MDA-MB-231, human TNBC cells, were treated with neem leaf extract (Neem) and eight, 1200 V/cm, 100 µs electric pulses (EP), and their viability and proteomic profiles were studied. With EP + Neem, a lower viability of 37% was observed after 24 h, compared to 85% in the neem-only samples, indicating the efficacy of the combinational treatment. The proteomics results indicated significant upregulation of 525 proteins and downregulation of 572 proteins, with a number of different pathways in each case. These include a diverse group of proteins, such as receptors, heat shock proteins, and many others. The upregulated TCA cycle and OXPHOS pathways and the downregulated DNA replication and ubiquitin-mediated proteolytic pathways were associated with effective cell death, demonstrating the potency of this treatment. Viability results reveal the efficacious anticancer effects of the EP + Neem combination, via growth inhibition, on TNBC cells. Proteomics studies could readily identify the effected protein pathways, and their corresponding genes, that are responsible for cell death. This represents a potential therapeutic strategy against TNBC when patients are refractory to standard treatments.
















Similar content being viewed by others
Availability of Data and Materials
All data sources could be available to readers on request.
References
Breast Cancer Overtakes Lung Cancer. Available from: https://www.reuters.com/article/us-health-cancer/breast-cancer-overtakes-lung-as-most-common-cancer-who-idUSKBN2A21AK. [ Last Accessed Feb 02 2021.]
Masuda, H., Baggerly, K. A., Wang, Y., Zhang, Y., Gonzalez-Angulo, A. M., Meric-Bernstam, F., ... & Ueno, N. T. (2013). Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clinical cancer research, 19(19), 5533-5540
Kosok, M., Alli-Shaik, A., Bay, B. H., & Gunaratne, J. (2020). Comprehensive proteomic characterization reveals subclass-specific molecular aberrations within triple-negative breast cancer. Iscience, 23(2), 100868.
Malla, R. R., Deepak, K. G. K., Merchant, N., & Dasari, V. R. (2020). Breast tumor microenvironment: Emerging target of therapeutic phytochemicals. Phytomedicine, 153227.
Lehmann, B. D., Bauer, J. A., Chen, X., Sanders, M. E., Chakravarthy, A. B., Shyr, Y., & Pietenpol, J. A. (2011). Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation, 121(7), 2750–2767.
Liu, N. Q., Stingl, C., Look, M. P., Smid, M., Braakman, R. B., De Marchi, T., ... & Umar, A. (2014). Comparative proteome analysis revealing an 11-protein signature for aggressive triple-negative breast cancer. JNCI: Journal of the National Cancer Institute, 106(2)
Lawrence, R. T., Perez, E. M., Hernández, D., Miller, C. P., Haas, K. M., Irie, H. Y., ... & Villén, J. (2015). The proteomic landscape of triple-negative breast cancer. Cell reports, 11(4), 630-644.
Miah, S., Banks, C. A., Adams, M. K., Florens, L., Lukong, K. E., & Washburn, M. P. (2017). Advancement of mass spectrometry-based proteomics technologies to explore triple negative breast cancer. Molecular BioSystems, 13(1), 42–55.
Hill, D. P., Harper, A., Malcolm, J., McAndrews, M. S., Mockus, S. M., Patterson, S. E., ... & Blake, J. A. (2019). Cisplatin-resistant triple-negative breast cancer subtypes: Multiple mechanisms of resistance. BMC cancer, 19(1), 1-13
Zou, W., Yang, Y., Zheng, R., Wang, Z., Zeng, H., Chen, Z., ... & Wang, J. (2020). Association of CD44 and CD24 phenotype with lymph node metastasis and survival in triple-negative breast cancer. International Journal of Clinical and Experimental Pathology, 13(5), 1008
Sharma, R., Kaushik, S., Shyam, H., Agarwal, S., & Balapure, A. K. (2017). Neem seed oil induces apoptosis in MCF-7 and MDA MB-231 human breast cancer cells. Asian Pacific journal of cancer prevention: APJCP, 18(8), 2135.
Sharma, C., Vas, A. J., Goala, P., Gheewala, T. M., Rizvi, T. A., & Hussain, A. (2014). Ethanolic neem (Azadirachta indica) leaf extract prevents growth of MCF-7 and HeLa cells and potentiates the therapeutic index of cisplatin. Journal of oncology, 2014, 321754.
Moga, M. A., Bălan, A., Anastasiu, C. V., Dimienescu, O. G., Neculoiu, C. D., & Gavriș, C. (2018). An overview on the anticancer activity of Azadirachta indica (Neem) in gynecological cancers. International journal of molecular sciences, 19(12), 3898. https://doi.org/10.3390/ijms1912389813
Othman, F., Motalleb, G., Peng, S. L. T., Rahmat, A., Basri, R., & Pei, C. P. (2012). Effect of neem leaf extract (Azadirachta indica) on c-Myc oncogene expression in 4T1 breast cancer cells of BALB/c mice. Cell Journal (Yakhteh), 14(1), 53.
Boopalan, T., Arumugam, A., Damodaran, C., & Rajkumar, L. (2012). The anticancer effect of 2’-3’-dehydrosalannol on triple-negative breast cancer cells. Anticancer research, 32(7), 2801–2806.
Marty, M., Sersa, G., Garbay, J. R., Gehl, J., Collins, C. G., Snoj, M., ... & Mir, L. M. (2006). Electrochemotherapy–An easy, highly effective and safe treatment of cutaneous and subcutaneous metastases: Results of ESOPE (European Standard Operating Procedures of Electrochemotherapy) study. European Journal of Cancer Supplements, 4(11), 3-13. doi:https://doi.org/10.1007/s11517-012-0991-8
Serša, G., Čemažar, M., & Miklavčič, D. (1995). Antitumor effectiveness of electrochemotherapy with cis-diamminedichloroplatinum (II) in mice. Cancer research, 55(15), 3450–3455.
Campana, L. G., Miklavčič, D., Bertino, G., Marconato, R., Valpione, S., Imarisio, I., ... & Sersa, G. (2019, April). Electrochemotherapy of superficial tumors–Current status:: Basic principles, operating procedures, shared indications, and emerging applications. In Seminars in oncology (Vol. 46, No. 2, pp. 173–191). WB Saunders. https://doi.org/10.1053/j.seminoncol.2019.04.002.
Clover, et al. Electrochemotherapy in the treatment of cutaneous malignancy: Outcomes and subgroup analysis from the cumulative results from the pan-European International Network for sharing practice in Electrochemotherapy database for 2482 lesions in 987 patients (2008-2019). European Journal of Cancer 2020, 138, pp. 30-40. doi: https://doi.org/10.1016/j.ejca.2020.06.020
Hansen, H. F., Bourke, M., Stigaard, T., Clover, J., Buckley, M., O’Riordain, M., ... & Gehl, J. (2020). Electrochemotherapy for colorectal cancer using endoscopic electroporation: a phase 1 clinical study. Endoscopy international open, 8(2), E124
Wichtowski, M., Murawa, D., Czarnecki, R., Piechocki, J., Nowecki, Z., & Witkiewicz, W. (2019). Electrochemotherapy in the treatment of breast cancer metastasis to the skin and subcutaneous tissue-multicenter experience. Oncology research and treatment, 42(1–2), 47–51.
How can proteomics boost the discovery of novel drugs and biomarkers? https://www.labiotech.eu. Acessed Jan 2021.
Mittal, L., Aryal, U. K., Camarillo, I. G., Ferreira, R. M., & Sundararajan, R. (2019). Quantitative proteomic analysis of enhanced cellular effects of electrochemotherapy with Cisplatin in triple-negative breast cancer cells. Scientific reports, 9(1), 1–16.
Mittal, L., Aryal, U. K., Camarillo, I. G., Raman, V., & Sundararajan, R. (2020). Effective electrochemotherapy with curcumin in MDA-MB-231-human, triple negative breast cancer cells: A global proteomics study. Bioelectrochemistry, 131, 107350. https://doi.org/10.1016/j.bioelechem.2019.107350
Montgomery, D. C., Peck, E. A., & Vining, G. G. (2012). Introduction to linear regression analysis (Vol. 821). John Wiley & Sons.
Sherman, B. T., & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols, 4(1), 44.
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., ... & Mering, C. V. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research, 47(D1), D607-D613.
Carmona-Saez, P., Chagoyen, M., Tirado, F., Carazo, J. M., & Pascual-Montano, A. (2007). GENECODIS: A web-based tool for finding significant concurrent annotations in gene lists. Genome biology, 8(1), 1–8.
Huang, D. W., Sherman, B. T., & Lempicki, R. A. (2009). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research, 37(1), 1–13.
Walczak, A., Gradzik, K., Kabzinski, J., Przybylowska-Sygut, K., & Majsterek, I. (2019). The role of the ER-induced UPR pathway and the efficacy of its inhibitors and inducers in the inhibition of tumor progression. Oxidative medicine and cellular longevity, 2019, 5729710.
Oslowski, C. M., & Urano, F. (2011). Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods in enzymology, 490, 71–92.
Boyer, A. S., Walter, D., & Sørensen, C. S. (2016). DNA replication and cancer: From dysfunctional replication origin activities to therapeutic opportunities. In Seminars in cancer biology, 37, 16–25. Academic Press.
Myung, J., Kim, K. B., & Crews, C. M. (2001). The ubiquitin-proteasome pathway and proteasome inhibitors. Medicinal research reviews, 21(4), 245–273.
Nuñez, G., Benedict, M. A., Hu, Y., & Inohara, N. (1998). Caspases: The proteases of the apoptotic pathway. Oncogene, 17(25), 3237–3245.
Elena-Real, C. A., Díaz-Quintana, A., González-Arzola, K., Velázquez-Campoy, A., Orzáez, M., López-Rivas, A., ... & Díaz-Moreno, I. (2018). Cytochrome c speeds up caspase cascade activation by blocking 14-3-3ε-dependent Apaf-1 inhibition. Cell death & disease, 9(3), 1-12
Benov, L. C., Antonov, P. A., & Ribarov, S. R. (1994). Oxidative damage of the membrane lipids after electroporation. General physiology and biophysics, 13, 85–85.
Gabriel, B., & Teissie, J. (1994). Generation of reactive-oxygen species induced by electropermeabilization of Chinese hamster ovary cells and their consequence on cell viability. European Journal of Biochemistry, 223(1), 25–33.
Biebl, M. M., & Buchner, J. (2019). Structure, function, and regulation of the Hsp90 machinery. Cold Spring Harbor perspectives in biology, 11(9), a034017.
Klimczak, M., Biecek, P., Zylicz, A., & Zylicz, M. (2019). Heat shock proteins create a signature to predict the clinical outcome in breast cancer. Scientific reports, 9(1), 1–15.
Zoppino, F. C., Guerrero-Gimenez, M. E., Castro, G. N., & Ciocca, D. R. (2018). Comprehensive transcriptomic analysis of heat shock proteins in the molecular subtypes of human breast cancer. BMC Cancer, 18(1), 1–17.
Pragatheiswar, G., Mittal, L., Camarillo, I. G., & Sundararajan, R. (2021). Analysis of pathways in triple-negative breast cancer cells treated with the combination of electrochemotherapy and cisplatin. Biointerface Research in Applied Chemistry, 11(5), 13453–13464. https://doi.org/10.33263/BRIAC115.1345313464
Acknowledgements
Jeya Shree T is very grateful to the 2019-2020 SERB Fellowship that enabled her to visit Purdue and do the experiments. All the mass spec experiments were performed at the Purdue Proteomics Facility, Bindley Bioscience Center. We are very grateful to its Director, Dr. Uma Aryal, for his kind help. The Q Exactive Orbitrap HF and UltiMate 3000 HPLC system used for LC-MS/MS analysis were purchased from funding provided by the Purdue Office of the Executive Vice President for Research and Partnership. The authors are grateful to Dr. Laura W. Bower’s lab for the use of a spectrophotometer, and Drs. Emily C. Dykhuizen’s and Ourania M. Andrisani’s labs for their qPCR machines. The authors are grateful to Poompavai and Lakshya Mittal for their help with sample collection and data analysis.
Author information
Authors and Affiliations
Contributions
Concept and design: Gowrisree V, R Sundararajan, IG Camarillo. Neem leaf collection and extract preparation: Gowrisree V. Viability and other Experiments: Jeya Shree T, IG Camarillo. Data Analysis: P Giri, Jeya Shree T, R Sundararajan. Manuscript: All.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Varadarajan, G., Thulasidas, J.S., Giri, P. et al. High-throughput, Label-free Proteomics Identifies Salient Proteins and Genes in MDA-MB-231 Cells Treated with Natural Neem-based Electrochemotherapy. Appl Biochem Biotechnol 194, 148–166 (2022). https://doi.org/10.1007/s12010-021-03787-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03787-3