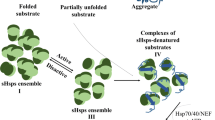
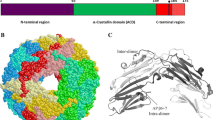
Abstract
Small heat shock proteins (sHSPs), often known as molecular chaperones, are most prevalent in nature. Under certain stress-induced conditions, these sHSPs act as an ATP-independent variation and thus prevent the inactivation of various non-native substrate proteins and their aggregation. They also assist other ATP-dependent chaperones in the refolding of these substrates. In the case of prokaryotes and lower eukaryotes, the chaperone functions of sHSPs can bind a wide range of cellular proteins but preferentially protect translation-related proteins and metabolic enzymes. Eukaryotes usually encode a larger number of sHSPs than those of prokaryotes. The chaperone functions of mammalian sHSPs are regulated by phosphorylation in cells and also by temperature. Their sHSPs have different sub-cellular compartments and cell/tissue specificity. The substrate proteins of mammalian sHSPs or eukaryotic sHSPs accordingly reflect their multi-cellular complexity. The sHSPs of animals play roles in different physiological processes as cell differentiation, apoptosis, and longevity. In this work, the characterization, location, tissue specificity, and functional diversity of sHSPs from seven different mammalian species with special emphasis on humans have been studied. Through this extensive work, a novel and significant attempt have been made to classify them based on their omnipresence, tissue specificity, localization, secondary structure, probable mutations, and evolutionary significance.




Similar content being viewed by others
Data Availability
The data are available upon request.
References
Heirbaut, M., Beelen, S., Strelkov, S. V., & Weeks, S. D. (2014). Dissecting the functional role of the N-terminal domain of the human small heat shock protein HSPB6. PLoS One, 9(8), e105892. https://doi.org/10.1371/journal.pone.0105892.
Ferns, G., Shams, S., & Shafi, S. (2006). Heat shock protein 27: its potential role in vascular disease. International Journal of Experimental Pathology, 87(4), 253–274. https://doi.org/10.1111/j.1365-2613.2006.00484.x.
Mogk, A., & Bukau, B. (2017). Role of sHSPs in organizing cytosolic protein aggregation and disaggregation. Cell Stress & Chaperones, 22(4), 493–502. https://doi.org/10.1007/s12192-017-0762-4.
Sugiyama, Y., Suzuki, A., Kishikawa, M., Akutsu, R., Hirose, T., & Waye, M. M. (2000). Muscle develops a specific form of small heat shock protein complex composed of MKBP/HSPB2 and HSPB3 during myogenic differentiation. The Journal of Biological Chemistry, 275(2), 1095–1104. https://doi.org/10.1074/jbc.275.2.1095.
Yoshida, K., Aki, T., Harada, K., Shama, K. M., Kamoda, Y., Suzuki, A., & Ohno, S. (1999). Translocation of HSP27 and MKBP in ischemic heart. Cell Structure and Function, 24(4), 181–185. https://doi.org/10.1247/csf.24.181.
Mymrikov, E., Seit-Nebi, A., & Gusev, N. (2011). Large potentials of small heat shock proteins. Physiological Reviews, 91(4), 1123–1159. https://doi.org/10.1152/physrev.00023.
Kriehuber, T., Rattei, T., Weinmaier, T., Bepperling, A., Haslbeck, M., & Buchner, J. (2010). Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology, 24(10), 3633–3642. https://doi.org/10.1096/fj.10-156992.
Taylor, R. P., & Benjamin, I. J. (2005). Small heat shock proteins: a new classification scheme in mammals. Journal of Molecular and Cellular Cardiology, 38(3), 433–444. https://doi.org/10.1016/j.yjmcc.2004.12.014.
Benjamin, I. J., & McMillan, D. R. (1998). Stress (heat shock) proteins: molecular chaperones in cardiovascular biology and disease. Circulation Research, 83(2), 117–132. https://doi.org/10.1161/01.res.83.2.117.
Lam, W. Y., Wing, S. K., Tsui, P. T., Law, Luk, S. C., Fung, K. P., & Lee, C. Y. (1996). Isolation and characterization of a human heart cDNA encoding a new member of the small heat shock protein family—HSPL27. Biochimica et Biophysica Acta, 1314(1-2), 120–124. https://doi.org/10.1016/s0167-4889(96)00121-8.
The UniProt Consortium. (2019). UniProt: the universal protein knowledgebase. Nucleic Acids Research, 47(D1), D506–D515. https://doi.org/10.1093/nar/gky1049.
Itaya, H., Oshita, K., Arakawa, K., & Tomita, M. (2013). GEMBASSY: an EMBOSS associated software package for comprehensive genome analyses. Source Code for Biology and Medicine, 8(1), 17. https://doi.org/10.1186/1751-0473-8-17.
Simossis, V. A., & Heringa, J. (2005). PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Research, 33(Web Server issue), W289–W294. https://doi.org/10.1093/nar/gki390.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673–4680. https://doi.org/10.1093/nar/22.22.4673.
El-Gebali, S., Mistry, J., Bateman, A., Eddy, R. S., Luciani, A., Potter, C. S., Qureshi, M., Richardson, J. L., Salazar, A. G., Smart, A., Sonnhammer, L. L. E., Hirsh, L., Paladin, L., Piovesan, D., Tosatto, E. C. S., & Finn, D. R. (2019). The Pfam protein families database in 2019.Nucleic. Acids Research, 47(D1), D427–D432. https://doi.org/10.1093/nar/gky995.
Narberhaus, F. (2002). Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiology and Molecular Biology Reviews: MMBR, 66(1), 64–93. https://doi.org/10.1128/mmbr.66.1.64-93.2002.
Kouza, M., Faraggi, E., Kolinski, A., & Kloczkowski, A. (2017). The GOR method of protein secondary structure prediction and its application as a protein aggregation prediction tool. Methods in Molecular Biology (Clifton, N.J.), 1484, 7–24. https://doi.org/10.1007/978-1-4939-6406-2_2.
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., Dufayard, J. F., Guindon, S., Lefort, V., Lescot, M., Claverie, J. M., & Gascuel, O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research, 36(Web Server issue), W465–W469. https://doi.org/10.1093/nar/gkn180.
Islamovic, E., Duncan, A., Bers, D. M., Gerthoffer, W. T., & Mestril, R. (2007). Importance of small heat shock protein 20 (hsp20) C-terminal extension in cardioprotection. Journal of Molecular and Cellular Cardiology, 42(4), 862–869. https://doi.org/10.1016/j.yjmcc.2007.01.002.
Boelens, W. C., Van Boekel, M. A., & De Jong, W. W. (1998). HspB3, the most deviating of the six known human small heat shock protein. Biochimica et Biophysica Acta, 1388(2), 513–516. https://doi.org/10.1016/s0167-4838(98)00215-5.
Sha, L., Hou, N., Zhang, M., Ma, Q., & Shi, C. (2019). High α B-crystallin and p53 co-expression is associated with poor prognosis in ovarian cancer. Bioscience Reports, 39(6), BSR20182407. https://doi.org/10.1042/BSR20182407.
Iwaki, A., Nagano, T., Nakagawa, M., Iwaki, T., & Fukumaki, Y. (1997). Identification and characterization of the gene encoding a new member of the alpha-crystallin/small HSP family, closely linked to the alpha-B crystallin gene in a head-to-head manner. Genomics, 45(2), 386–394. https://doi.org/10.1006/geno.1997.4956.
Arrigo, A. P., Simon, S., Gibert, B., Kretz-Remy, C., Nivon, M., Czekalla, A., Guillet, D., Moulin, M., Diaz-Latoud, C., & Vicart, P. (2007). Hsp27 (HspB1) and alphaB-crystallin (HspB5) as therapeutic targets. FEBS Letters, 581(19), 3665–3674. https://doi.org/10.1016/j.febslet.2007.04.033.
Suzuki, A., Sugiyama, Y., Hayashi, Y., Nyu-i, N., Yoshida, M., & Nonaka, I. (1998). A novel member of the small heat shock protein family, binds and activates the myotonic dystrophy protein kinase. The Journal of Cell Biology, 140(5), 1113–1124. https://doi.org/10.1083/jcb.140.5.1113.
McGuffin, L., Bryson, K., & Jones, D. (2000). The PSIPRED protein structure prediction server. Bioinformatics (Oxford, England), 16(4), 404–405. https://doi.org/10.1093/bioinformatics/16.4.404.
Gastmann, O., Burfeind, P., Gunther, E., Hameister, H., Szpirer, C., & Hoyer-Fender, S. (1993). Sequence, expression, and chromosomal assignment of a human sperm outer dense fiber gene. Molecular Reproduction and Development, 36(4), 407–418. https://doi.org/10.1002/mrd.1080360402.
Waters, E. R., & Rioflorido, I. (2007). Evolutionary analysis of the small heat shock proteins in five complete algal genomes. Journal of Molecular Evolution, 65(2), 162–174. https://doi.org/10.1007/s00239-006-0223-7.
Fan, G. C., Chu, G., Mitton, B., Song, Q., Yuan, Q., & Kranias, E. G. (2004). Small heat-shock protein Hsp20 phosphorylation inhibits β-agonist-induced cardiac apoptosis. Circulation Research, 94(11), 1474–1482. https://doi.org/10.1161/01.res.0000129179.66631.00.
Basha, E., Jones, C., Wysocki, V., & Vierling, E. (2010). Mechanistic differences between two conserved classes of small heat shock proteins found in the plant cytosol. The Journal of Biological Chemistry, 285(15), 11489–11497. https://doi.org/10.1074/jbc.M109.074088.
Siddique, M., Gernhard, S., Koskull-Doring, P., Vierling, E., & Scharf, K. D. (2008). The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress & Chaperones, 13(2), 183–197. https://doi.org/10.1007/s12192-008-0032-6.
van Heijst, J. W., Niessen, H. W., Musters, R. J., van Hinsbergh, V. W., Hoekman, K., & Schalkwijk, C. G. (2006). Argpyrimidine-modified heat shock protein 27 in human non-small cell lung cancer: a possible mechanism for evasion of apoptosis. Cancer Letters, 241(2), 309–319. https://doi.org/10.1016/j.canlet.2005.10.042.
Nicolaou, P., Knöll, R., Haghighi, K., Guo-Chang, F., Dorn, G. W., Hasenfu, G., & Kranias, E. G. (2008). Human mutation in the anti-apoptotic heat shock protein 20 abrogates its cardioprotective effects. The Journal of Biological Chemistry, 283(48), 33465–33471. https://doi.org/10.1074/jbc.M802307200.
Sigrist, C. J., Cerutti, L., Hulo, N., Gattiker, A., Falquet, L., Pagni, M., Bairoch, A., & Bucher, P. (2002). PROSITE: a documented database using patterns and profiles as motif descriptors. Briefings in Bioinformatics, 3(3), 265–274. https://doi.org/10.1093/bib/3.3.265.
Chu, G., Egnaczyk, G. F., Zhao, W., Jo, S. H., Fan, G. C., Maggio, J. E., Xiao, R. P., & Kranias, E. G. (2004). Phosphoproteome analysis of cardiomyocytes subjected to beta-adrenergic stimulation: identification and characterization of a cardiac heat shock protein p20. Circulation Research, 94(2), 184–193. https://doi.org/10.1161/01.RES.0000107198.90218.21.
Pandey, B., Kaur, A., Gupta, O. P., Sharma, I., & Sharma, P. (2000). Identification of HSP20 gene family in wheat and barley and their differential expression profiling under heat stress. Applied Biochemistry and Biotechnology, 175(5), 2427–2446. https://doi.org/10.1007/s12010-014-1420-2.
Zhu, Y. H., Ma, T. M., & Wang, X. (2005). Gene transfer of heat-shock protein 20 protects against ischemia/reperfusion injury in rat hearts. Acta Pharmacologica Sinica, 26(10), 1193–1200. https://doi.org/10.1111/j.1745-7254.2005.00139.x.
Kappé, G., Franck, E., Verschuure, P., Boelens, W. C., Leunissen, J. A., & de Jong, W. W. (2003). The human genome encodes 10 alpha-crystallin-related small heat shock proteins: HspB1-10. Cell Stress & Chaperones, 8(1), 53–61. https://doi.org/10.1379/1466-1268(2003)8<53:thgecs>2.0.co;2.
Acknowledgments
The authors acknowledge the help from the DBT-funded BIF (Bioinformatics facility center), University of Kalyani, West Bengal are also acknowledged for furnishing all the necessary infrastructural facilities to pursue the whole study.
Funding
The research work is supported by the Department of Science and Technology and Biotechnology of West Bengal Govt. who rendered required financial support to carry on with the research works through R&D project SA. No./ST/P/S&T/1-G14/2018. Financial support was also provided by UGC-SAP-DSR-II and DST-PURSE-2.
Author information
Authors and Affiliations
Contributions
AB and RDG devised the project. SM performed the experimentations. All the authors wrote the manuscript and agreed to submit the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
Not applicable
Consent to Participate
Not applicable
Consent to Publish
All authors agreed to submit the manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mitra, S., Bagchi, A. & Dasgupta, R. Elucidation of Diverse Physico-Chemical Parameters in Mammalian Small Heat Shock Proteins: A Comprehensive Classification and Structural and Functional Exploration Using In Silico Approach. Appl Biochem Biotechnol 193, 1836–1852 (2021). https://doi.org/10.1007/s12010-021-03497-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03497-w