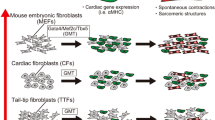
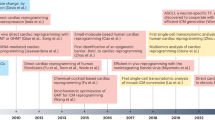
Abstract
Purpose of Review
The high global incidence of heart disease drives the need for methods of mending damaged hearts. Direct reprogramming of cardiac fibroblasts into cardiomyocyte-like cells (called iCMs) has been successful in the creation of new muscle cells, in the repair of hearts post-myocardial injury, and therefore has great promise for the clinic. The purpose of this paper is to review and highlight the approaches for and underlying molecular mechanisms of direct cardiac reprogramming.
Recent Findings
Single-cell genomics and mechanistic studies have elucidated the stepwise transition of fibroblasts to iCMs as well as the molecular roadblocks that hinder reprogramming.
Summary
Cardiac fibroblasts are able to be directly reprogrammed, in vitro and in vivo, into induced cardiomyocyte-like cells by the ectopic expression of a combination of transcription factors, microRNAs or small molecules. Recent works have illustrated methods that improve the efficiency of iCM generation and delivery of reprogramming cocktails as well as have revealed the molecular networks governing the reprogramming process. Current studies have also begun to identify and address the additional hurdles in human iCM reprogramming.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y, et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature. 2016;538(7625):388–91.
Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109(27):E1848–57.
Zhou Y, Wang L, Liu Z, Alimohamadi S, Yin C, Liu J, et al. Comparative gene expression analyses reveal distinct molecular signatures between differentially reprogrammed cardiomyocytes. Cell Rep. 2017;20(13):3014–24.
Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–86.
Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485(7400):593–8.
Song K, Nam Y-J, Luo X, Qi X, Tan W, Huang GN, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485(7400):599–604.
Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53(3):323–32.
Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110(11):1465–73.
Jayawardena TM, Finch EA, Zhang L, Zhang H, Hodgkinson CP, Pratt RE, et al. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ Res. 2015;116(3):418–24.
Cao N, Huang Y, Zheng J, Spencer CI, Zhang Y, Fu J-D, et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science. 2016;352(6290):1216–20.
Zhou H, Dickson ME, Kim MS, Bassel-Duby R, Olson EN. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci U S A. 2015;112(38):11864–9.
Hoque A, Sivakumaran P, Bond ST, Ling NXY, Kong AM, Scott JW, et al. Mitochondrial fission protein Drp1 inhibition promotes cardiac mesodermal differentiation of human pluripotent stem cells. Cell Death Discov. 2018;4:39.
Sutcliffe MD, Tan PM, Fernandez-Perez A, Nam Y-J, Munshi NV, Saucerman JJ. High content analysis identifies unique morphological features of reprogrammed cardiomyocytes. Sci Rep. 2018;8(1):1258.
Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106.
Keepers B, Liu J, Qian L. What's in a cardiomyocyte - And how do we make one through reprogramming?. Biochim Biophys Acta Mol Cell Res. 2020;1867(3):118464. https://doi.org/10.1016/j.bbamcr.2019.03.011.
Christoforou N, Chellappan M, Adler AF, Kirkton RD, Wu T, Addis RC, et al. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One. 2013;8(5):e63577.
• Zhou Y, Liu Z, Welch JD, Gao X, Wang L, Garbutt T, et al. Single-cell transcriptomic analyses of cell fate transitions during human cardiac reprogramming. Cell Stem Cell. 2019;25(1):149–164.e9. This study provides the first molecular roadmap of human cardiac reprogramming at single cell level, highlights the possible regulatory pathways and molecules.
•• Liu Z, Wang L, Welch JD, Ma H, Zhou Y, Vaseghi HR, et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature. 2017;551(7678):100–4. This work for the first time provides the trajectory of how a murine fibroblast is directly converted into an iCM using single cell transcriptomics.
Welch JD, Hartemink AJ, Prins JF. SLICER: inferring branched, nonlinear cellular trajectories from single cell RNA-seq data. Genome Biol. 2016;17(1):106.
La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. 2018;560(7719):494–8.
• Stone NR, Gifford CA, Thomas R, Pratt KJB, Samse-Knapp K, Mohamed TMA, et al. Context-specific transcription factor functions regulate Epigenomic and transcriptional dynamics during cardiac reprogramming. Cell Stem Cell. 2019;25(1):87–102.e9. This study demonstrates the early transcriptome and epigenome shift guided by MGT during muine cardiac reprogramming.
Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502.
Chen JX, Krane M, Deutsch M-A, Wang L, Rav-Acha M, Gregoire S, et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ Res. 2012;111(1):50–5.
Mahmoudi S, Mancini E, Xu L, Moore A, Jahanbani F, Hebestreit K, et al. Heterogeneity in old fibroblasts is linked to variability in reprogramming and wound healing. Nature. 2019;574(7779):553–8.
Wang L, Liu Z, Yin C, Asfour H, Chen O, Li Y, et al. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ Res. 2015;116(2):237–44.
Ma H, Wang L, Yin C, Liu J, Qian L. In vivo cardiac reprogramming using an optimal single polycistronic construct. Cardiovasc Res. 2015;108(2):217–9.
Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33(14):1565–81.
Zhou Y, Wang L, Vaseghi HR, Liu Z, Lu R, Alimohamadi S, et al. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell. 2016;18(3):382–95.
Liu L, Lei I, Karatas H, Li Y, Wang L, Gnatovskiy L, et al. Targeting Mll1 H3K4 methyltransferase activity to guide cardiac lineage specific reprogramming of fibroblasts. Cell Discov. 2016;2:16036.
Mohamed TMA, Stone NR, Berry EC, Radzinsky E, Huang Y, Pratt K, et al. Chemical enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation. 2017;135(10):978–95.
Miyamoto K, Akiyama M, Tamura F, Isomi M, Yamakawa H, Sadahiro T, et al. Direct in vivo reprogramming with sendai virus vectors improves cardiac function after myocardial infarction. Cell Stem Cell. 2018;22(1):91–103.e5.
Mathison M, Singh VP, Chiuchiolo MJ, Sanagasetti D, Mao Y, Patel VB, et al. In situ reprogramming to transdifferentiate fibroblasts into cardiomyocytes using adenoviral vectors: Implications for clinical myocardial regeneration. J Thorac Cardiovasc Surg. 2017;153(2):329–339.e3.
Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O’Rourke R, et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun. 2015;6:8243.
Mathison M, Singh VP, Sanagasetti D, Yang L, Pinnamaneni JP, Yang J, et al. Cardiac reprogramming factor Gata4 reduces postinfarct cardiac fibrosis through direct repression of the profibrotic mediator snail. J Thorac Cardiovasc Surg. 2017;154(5):1601–1610.e3.
• Hashimoto H, Wang Z, Garry GA, Malladi VS, Botten GA, Ye W, et al. Cardiac reprogramming factors synergistically activate genome-wide cardiogenic stage-specific enhancers. Cell Stem Cell. 2019;25(1):69–86.e5. This work illustrates the role of each reprogramming factor in instructing murine iCM conversion.
Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci U S A. 2013;110(31):12667–72.
Fu J-D, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports. 2013;1(3):235–47.
Nam Y-J, Song K, Luo X, Daniel E, Lambeth K, West K, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A. 2013;110(14):5588–93.
Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–81.
Singh VP, Mathison M, Patel V, et al. MiR-590 Promotes Transdifferentiation of Porcine and Human Fibroblasts Toward a Cardiomyocyte-Like Fate by Directly Repressing Specificity Protein 1. J Am Heart Assoc. 2016;5(11):e003922. https://doi.org/10.1161/JAHA.116.003922.
Patel V, Singh VP, Pinnamaneni JP, Sanagasetti D, Olive J, Mathison M, et al. p63 Silencing induces reprogramming of cardiac fibroblasts into cardiomyocyte-like cells. J Thorac Cardiovasc Surg. 2018;156(2):556–565.e1.
Mathison M, Gersch RP, Nasser A, Lilo S, Korman M, Fourman M, et al. In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J Am Heart Assoc. 2012;1(6):e005652.
Wang L, Huang P, Near D, et al. Isoform Specific Effects of Mef2C during Direct Cardiac Reprogramming. Cells. 2020;9(2):268. https://doi.org/10.3390/cells9020268.
Lee S, Lee DH, Park B-W, Kim R, Hoang AD, Woo S-K, et al. In vivo transduction of ETV2 improves cardiac function and induces vascular regeneration following myocardial infarction. Exp Mol Med. 2019;51(2):13.
Shadrin IY, Allen BW, Qian Y, Jackman CP, Carlson AL, Juhas ME, et al. Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat Commun. 2017;8(1):1825.
Gao L, Gregorich ZR, Zhu W, Mattapally S, Oduk Y, Lou X, et al. Large cardiac muscle patches engineered from human induced-pluripotent stem cell-derived cardiac cells improve recovery from myocardial infarction in swine. Circulation. 2018;137(16):1712–30.
Chen J, Guo Z, Tian H, Chen X. Production and clinical development of nanoparticles for gene delivery. Mol Ther Methods Clin Dev. 2016;3:16023.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–83.
Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160(1–2):339–50.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Gregory Farber and Li Qian declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Regenerative Medicine
Rights and permissions
About this article
Cite this article
Farber, G., Qian, L. Reprogramming of Non-myocytes into Cardiomyocyte-like Cells: Challenges and Opportunities. Curr Cardiol Rep 22, 54 (2020). https://doi.org/10.1007/s11886-020-01322-0
Published:
DOI: https://doi.org/10.1007/s11886-020-01322-0