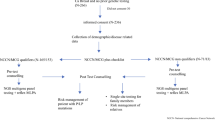
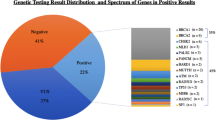
Abstract
Background
Breast cancer is genetically heterogeneous, and parellel multi-gene sequencing is the most cost- and time-efficient manner to investigate breast cancer predisposition. Numerous multi-gene panels (MGPs) are commercially available, but many include genes with weak/unproven associaton with breast cancer, or with predisposition to cancer of other types. This study investigates the utility of a custom-designed multi-gene panel in an Irish cohort with breast cancer.
Methods
A custom panel comprising 83 genes offered by 19 clinical “breast cancer predisposition” MGPs was designed and applied to germline DNA from 91 patients with breast cancer and 77 unaffected ethnicially matched controls. Variants were identified and classified using a custom pipeline.
Results
Nineteen loss-of-function (LOF) and 334 missense variants were identified. After removing common and/or benign variants, 15 LOF and 30 missense variants were analysed. Variants in known breast cancer susceptibility genes were identified, including in BRCA1 and ATM in cases, and in NF1 and CHEK2 in controls. Most variants identified were in genes associated with predisposition to cancers other than breast cancer (BRIP1, RAD50, MUTYH, and mismatch repair genes), or in genes with unknown or unproven association with cancer.
Conclusion
Using multi-gene panels enables rapid, cost-effective identification of individuals with high-risk cancer predisposition syndromes. However, this approach also leads to an increased amount of uncertain results. Clinical management of individuals with particular genetic variants in the absence of a matching phenotype/family history is challenging. Further population and functional evidence is required to fully elucidate the clinical relevance of variants in genes of uncertain significance.



Similar content being viewed by others
Change history
18 February 2020
The originally published version of this article contained typesetting errors in Figs.��2 and 3 legends. The correct figure legends are presented here. The original article has been corrected.
Abbreviations
- MAF:
-
Minor allele frequency
- NFE:
-
Non-Finnish European
- GWAS:
-
Genome-wide assocation study
- NGS:
-
Next-generation sequencing
- MGP:
-
Multi-gene panel
- VUS:
-
Variant of uncertain significance
- GATK:
-
Genome Analysis Toolkit
- LOF:
-
Loss-of-function
- SNP:
-
Single nucleotide polymorphism
- PCA:
-
Principle component analysis
- VUS:
-
Variant(s) of uncertain significance
- CRC:
-
Colorectal cancer
- LS:
-
Lynch syndrome
- LD:
-
Linkage disequilibrium
- GUS:
-
Gene(s) of uncertain significance
- InSiGHT:
-
The International Society for Gastrointestinal Hereditary Tumours
- ENIGMA:
-
Evidence-based Network for the Interpretation of Germline Mutant Alleles
References
Peto J, Mack TM (2000) High constant incidence in twins and other relatives of women with breast cancer. Nat Genet 26(4):411–414
Newman B, Austin MA, Lee M, King MC (1988) Inheritance of human breast cancer: evidence for autosomal dominant transmission in high-risk families. Proc Natl Acad Sci U S A 85(9):3044–3048
Lynch HT, Silva E, Snyder C, Lynch JF (2008) Hereditary breast cancer: part I. Diagnosing hereditary breast cancer syndromes. Breast J 14(1):3–13
Ghoussaini M, Pharoah PD (2009) Polygenic susceptibility to breast cancer: current state-of-the-art. Future Oncol 5(5):689–701
Easton DF, Pharoah PD, Antoniou AC et al (2015) Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 372(23):2243–2257
Nelson HD et al. (2013) U.S. preventive services task force evidence syntheses, formerly systematic evidence reviews, in risk assessment, genetic counseling, and genetic testing for BRCA-related cancer: systematic review to update the U.S. Preventive Services Task Force Recommendation. Agency for Healthcare Research and Quality (US): Rockville (MD)
Thompson ER, Rowley SM, Li N et al (2016) Panel testing for familial breast cancer: calibrating the tension between research and clinical care. J Clin Oncol 34(13):1455–1459
Maxwell KN, Domchek SM, Nathanson KL, Robson ME (2016) Population frequency of germline BRCA1/2 mutations. J Clin Oncol 34(34):4183–4185
Nelson HD, Pappas M, Zakher B et al (2014) Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: a systematic review to update the U.S. Preventive Services Task Force recommendation. Ann Intern Med 160(4):255–266
Copson ER et al (2018) Germline BRCA mutation and outcome in young-onset breast cancer (POSH): a prospective cohort study. Lancet Oncol 19(2):169–180
Kuchenbaecker KB, Hopper JL, Barnes DR et al (2017) Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. Jama 317(23):2402–2416
Antoniou A, Pharoah PD, Narod S et al (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72(5):1117–1130
Chen S, Parmigiani G (2007) Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol 25(11):1329–1333
Tan MH, Mester JL, Ngeow J et al (2012) Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res 18(2):400–407
Schon K, Tischkowitz M (2018) Clinical implications of germline mutations in breast cancer: TP53. Breast Cancer Res Treat 167(2):417–423
Hearle N, Schumacher V, Menko FH et al (2006) Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res 12(10):3209–3215
Kaurah P, MacMillan A, Boyd N et al (2007) Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. Jama 297(21):2360–2372
Rahman N, Seal S, Thompson D et al (2007) PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 39(2):165–167
Sharif S et al (2007) Women with neurofibromatosis 1 are at a moderately increased risk of developing breast cancer and should be considered for early screening. J Med Genet 44(8):481–484
Meijers-Heijboer H, van den Ouweland A, Klijn J et al (2002) Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 31(1):55–59
Michailidou K et al (2017) Association analysis identifies 65 new breast cancer risk loci. Nature 551:92
Buniello A, MacArthur J, Cerezo M et al (2019) The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47(D1):D1005–d1012
ASSOC. FOR MOLECULAR PATHOLOGY v. MYRIAD GENETICS, INC. 2012, Supreme Court
TCGA Research Network. Available from: http://cancergenome.nih.gov/. Accessed 22 Jan 2020
van Marcke C et al (2016) Routine use of gene panel testing in hereditary breast cancer should be performed with caution. Crit Rev Oncol Hematol 108:33–39
Lynce F and C Isaacs (2016) How far do we go with genetic evaluation? Gene, panel, and tumor testing. American Society of Clinical Oncology educational book. American Society of Clinical Oncology. Annual Meeting. 35: p. e72–8
O'Leary E et al (2017) Expanded gene panel use for women with breast cancer: identification and intervention beyond breast cancer risk. Ann Surg Oncol 24(10):3060–3066
Buys SS, Sandbach JF, Gammon A et al (2017) A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 123(10):1721–1730
Lerner-Ellis J et al (2015) Genetic risk assessment and prevention: the role of genetic testing panels in breast cancer. Expert Rev Anticancer Ther 15(11):1315–1326
Kapoor NS et al (2015) Multigene panel testing detects equal rates of pathogenic BRCA1/2 mutations and has a higher diagnostic yield compared to limited BRCA1/2 analysis alone in patients at risk for hereditary breast cancer. Ann Surg Oncol 22(10):3282–3288
Crawford B et al (2017) Multi-gene panel testing for hereditary cancer predisposition in unsolved high-risk breast and ovarian cancer patients. Breast Cancer Res Treat 163(2):383–390
Manchanda R, Patel S, Gordeev VS et al (2018) Cost-effectiveness of population-based BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2 mutation testing in unselected general population women. J Natl Cancer Inst 110(7):714–725
Asphaug L, Melberg HO (2019) The cost-effectiveness of multigene panel testing for hereditary breast and ovarian cancer in Norway. MDM Policy Pract 4(1):2381468318821103
Li Y, Arellano AR, Bare LA et al (2017) A multigene test could cost-effectively help extend life expectancy for women at risk of hereditary breast cancer. Value Health 20(4):547–555
Wentzensen N, Berg CD (2018) Population testing for high penetrance genes: are we there yet? J Natl Cancer Inst 110(7):687–689
Evans DG, Harkness EF, Plaskocinska I et al (2017) Pathology update to the Manchester scoring system based on testing in over 4000 families. J Med Genet 54(10):674–681
McVeigh TP et al (2014) Familial breast cancer genetic testing in the west of Ireland. Ir J Med Sci 183(2):199–206
Collaborative Group on Hormonal Factors in Breast Cancer (2001) Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet 358(9291):1389–1399
Qiagen. Gentra Puregene Blood Kit. 2018 [cited 2018 July 4]; Available from: https://www.qiagen.com/us/shop/sample-technologies/dna/genomic-dna/gentra-puregene-blood-kit/#productdetails
Dnagenotek. DNA Extraction and Purification (prepIT-L2P | PT-L2P). 2018 [cited 2018 July 4]; Available from: https://www.dnagenotek.com/ROW/products/reagents-preparation/prepIT/PT-L2P.html
Andrews, S., FastQC: a quality control tool for high throughput sequence data. 2010
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997
Broad, I Picard toolkit (2018) Available from: http://broadinstitute.github.io/picard/. Accessed 22 Jan 2020
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303
DePristo MA et al (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43(5):491–498
Van der Auwera GA et al (2013) From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–11.1033
The Genomes Project C (2015) A global reference for human genetic variation. Nature 526:68
Cingolani P, Platts A, Wang le L et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2):80–92
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164–e164
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F (2016) The Ensembl variant effect predictor. Genome Biol 17(1):122
Shihab HA et al (2014) Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Human Genomics 8(1):11
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11(4):361–362
Reva B, Antipin Y, Sander C (2011) Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39(17):e118
Adzhubei IA, Schmidt S, Peshkin L et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249
Rentzsch P et al (2019) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47(D1):D886–d894
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42(Database issue):D980–D985
Couch FJ, Shimelis H, Hu C, Hart SN, Polley EC, Na J, Hallberg E, Moore R, Thomas A, Lilyquist J, Feng B, McFarland R, Pesaran T, Huether R, LaDuca H, Chao EC, Goldgar DE, Dolinsky JS (2017) Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncology 3(9):1190–1196
Lek M, Karczewski KJ, Minikel EV et al (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291
Karczewski KJ et al (2019) Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv:531210
National Comprehensive Cancer Network (2019) Guidelines for genetic/familial risk assessment: breast and ovarian. Version 3. 2019-January 18, 2019. Available from: https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf
Lee AJ et al (2016) Incorporating truncating variants in PALB2, CHEK2, and ATM into the BOADICEA breast cancer risk model. Genet Med 18(12):1190–1198
Pharoah PD, Guilford P, Caldas C (2001) Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology 121(6):1348–1353
Rich TA, Woodson AH, Litton J, Arun B (2015) Hereditary breast cancer syndromes and genetic testing. J Surg Oncol 111(1):66–80
Kamihara J, Rana HQ, Garber JE (2014) Germline TP53 mutations and the changing landscape of Li-Fraumeni syndrome. Hum Mutat 35(6):654–662
Southey MC, Teo ZL, Winship I (2013) PALB2 and breast cancer: ready for clinical translation! Appl Clin Genet 6:43–52
Antoniou AC, Casadei S, Heikkinen T et al (2014) Breast-cancer risk in families with mutations in PALB2. N Engl J Med 371(6):497–506
Southey MC, Teo ZL, Dowty JG et al (2010) A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res 12(6):R109–R109
Southey MC, Winship I, Nguyen-Dumont T (2016) PALB2: research reaching to clinical outcomes for women with breast cancer. Hereditary Cancer in Clinical Practice 14(1):9
Tavtigian SV et al (2009) Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. Am J Hum Genet 85(4):427–446
Schutte M, Seal S, Barfoot R et al (2003) Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am J Hum Genet 72(4):1023–1028
Nevanlinna H, Bartek J (2006) The CHEK2 gene and inherited breast cancer susceptibility. Oncogene 25(43):5912–5919
Schmidt MK, Hogervorst F, van Hien R et al (2016) Age- and tumor subtype-specific breast cancer risk estimates for CHEK2*1100delC carriers. J Clin Oncol 34(23):2750–2760
Vahteristo P, Bartkova J, Eerola H, Syrjäkoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomäki K, Heikkilä P, Holli K, Blomqvist C, Bartek J, Kallioniemi OP, Nevanlinna H (2002) A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet 71(2):432–438
Evans DG et al (2010) Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am J Med Genet A 152a(2):327–332
Seminog OO, Goldacre MJ (2015) Age-specific risk of breast cancer in women with neurofibromatosis type 1. Br J Cancer 112(9):1546–1548
Howell SJ, Hockenhull K, Salih Z, Evans DG (2017) Increased risk of breast cancer in neurofibromatosis type 1: current insights. Breast Cancer (Dove Med Press) 9:531–536
Taylor A, Brady AF, Frayling IM, Hanson H, Tischkowitz M, Turnbull C, Side L, UK Cancer Genetics Group (UK-CGG) (2018) Consensus for genes to be included on cancer panel tests offered by UK genetics services: guidelines of the UK cancer genetics group. J Med Genet 55(6):372–377
Office of the Associate Director for Science (OADS), Genomics & Precision Health, Centers for Disease Control and Prevention (2010) ACCE model process for evaluating genetic tests. Available from www.cdc.gov/genomics/gtesting/acce/. Accessed 22 Jan 2020
PHG Foundation. The personalised medicine technology landscape. [September 5, 2018 April 15, 2019]; Available from: http://www.phgfoundation.org/report/personalised-medicine-technology-landscape
McVeigh TP et al (2019) Managing uncertainty in inherited cardiac pathologies-an international multidisciplinary survey. Eur J Hum Genet
Sekine M, Nagata H, Tsuji S et al (2001) Mutational analysis of BRCA1 and BRCA2 and clinicopathologic analysis of ovarian cancer in 82 ovarian cancer families: two common founder mutations of BRCA1 in Japanese population. Clin Cancer Res 7(10):3144–3150
Abkevich V, Zharkikh A, Deffenbaugh AM, Frank D, Chen Y, Shattuck D, Skolnick MH, Gutin A, Tavtigian SV (2004) Analysis of missense variation in human BRCA1 in the context of interspecific sequence variation. J Med Genet 41(7):492–507
O'Donnell M, Axilbund J, Euhus DM (2018) In: Bland KI et al (eds) 17 - Breast cancer genetics: syndromes, genes, pathology, counseling, testing, and treatment, in the breast, 5th edn. Elsevier, pp 237–249.e5
Rousset-Jablonski C, Gompel A (2017) Screening for familial cancer risk: focus on breast cancer. Maturitas 105:69–77
Sato K, Koyasu M, Nomura S, Sato Y, Kita M, Ashihara Y, Adachi Y, Ohno S, Iwase T, Kitagawa D, Nakashima E, Yoshida R, Miki Y, Arai M (2017) Mutation status of RAD51C, PALB2 and BRIP1 in 100 Japanese familial breast cancer cases without BRCA1 and BRCA2 mutations. Cancer Sci 108(11):2287–2294
Easton DF, Lesueur F, Decker B et al (2016) No evidence that protein truncating variants in BRIP1 are associated with breast cancer risk: implications for gene panel testing. J Med Genet 53(5):298–309
Solyom S, Pylkas K, Winqvist R (2010) Screening for large genomic rearrangements of the BRIP1 and CHK1 genes in Finnish breast cancer families. Familial Cancer 9(4):537–540
Ameziane N, van den Ouweland A, Adank MA, Vijzelaar RN, Errami A, Dorsman JC, Joenje H, Meijers-Heijboer H, Waisfisz Q (2009) Lack of large genomic deletions in BRIP1, PALB2, and FANCD2 genes in BRCA1/2 negative familial breast cancer. Breast Cancer Res Treat 118(3):651–653
Theodoratou E, Campbell H, Tenesa A et al (2010) A large-scale meta-analysis to refine colorectal cancer risk estimates associated with MUTYH variants. Br J Cancer 103(12):1875–1884
Nielsen M, Franken PF, Reinards TH et al (2005) Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYH associated polyposis coli (MAP). J Med Genet 42(9):e54
Vogt S et al (2009) Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology 137(6):1976–85.e1–10
Wasielewski M, Out AA, Vermeulen J, Nielsen M, van den Ouweland A, Tops CM, Wijnen JT, Vasen HF, Weiss MM, Klijn JG, Devilee P, Hes FJ, Schutte M (2010) Increased MUTYH mutation frequency among Dutch families with breast cancer and colorectal cancer. Breast Cancer Res Treat 124(3):635–641
Out AA, Wasielewski M, Huijts PE, van Minderhout I, Houwing-Duistermaat JJ, Tops CM, Nielsen M, Seynaeve C, Wijnen JT, Breuning MH, van Asperen C, Schutte M, Hes FJ, Devilee P (2012) MUTYH gene variants and breast cancer in a Dutch case–control study. Breast Cancer Res Treat 134(1):219–227
Al-Tassan N et al (2002) Inherited variants of MYH associated with somatic G:C-->T: a mutations in colorectal tumors. Nat Genet 30(2):227–232
Cleary SP, Cotterchio M, Jenkins MA, Kim H, Bristow R, Green R, Haile R, Hopper JL, LeMarchand L, Lindor N, Parfrey P, Potter J, Younghusband B, Gallinger S (2009) Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology 136(4):1251–1260
McVeigh TP et al (2016) MUTYH-associated polyposis: the Irish experience>. Ir Med J 109(10):485
Roberts ME et al (2018) MSH6 and PMS2 germ-line pathogenic variants implicated in Lynch syndrome are associated with breast cancer. Genet Med
Goldberg M et al (2017) Association between the Lynch syndrome gene MSH2 and breast cancer susceptibility in a Canadian familial cancer registry. J Med Genet 54(11):742–746
Therkildsen C, Ladelund S, Smith-Hansen L, Lindberg LJ, Nilbert M (2017) Towards gene- and gender-based risk estimates in Lynch syndrome; age-specific incidences for 13 extra-colorectal cancer types. Br J Cancer 117(11):1702–1710
Moller P et al (2018) Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the prospective Lynch syndrome database. Gut 67(7):1306–1316
Muller A et al (2002) Exclusion of breast cancer as an integral tumor of hereditary nonpolyposis colorectal cancer. Cancer Res 62(4):1014–1019
de Leeuw WJ et al Correspondence re: A. Muller et al., Exclusion of breast cancer as an integral tumor of hereditary nonpolyposis colorectal cancer. Cancer Res. 62:1014–1019 2002. Cancer Res, 2003 63(5): p. 1148–9
Hampel H, Hall MJ (2018) Hereditary aspects of colorectal cancer: mismatch repair genes drive Lynch syndrome. J Adv Pract Oncol 9(3):311–315
Surtees JA, Alani E (2006) Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J Mol Biol 360(3):523–536
Palombo F et al (1996) hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol 6(9):1181–1184
Miao HK, Chen LP, Cai DP, Kong WJ, Xiao L, Lin J (2015) MSH3 rs26279 polymorphism increases cancer risk: a meta-analysis. Int J Clin Exp Pathol 8(9):11060–11067
Berndt SI, Platz EA, Fallin MD, Thuita LW, Hoffman SC, Helzlsouer KJ (2007) Mismatch repair polymorphisms and the risk of colorectal cancer. Int J Cancer 120(7):1548–1554
Jafary F, Salehi M, Sedghi M, Nouri N, Jafary F, Sadeghi F, Motamedi S, Talebi M (2012) Association between mismatch repair gene MSH3 codons 1036 and 222 polymorphisms and sporadic prostate cancer in the Iranian population. Asian Pac J Cancer Prev 13(12):6055–6057
Hirata H, Hinoda Y, Kawamoto K, Kikuno N, Suehiro Y, Okayama N, Tanaka Y, Dahiya R (2008) Mismatch repair gene MSH3 polymorphism is associated with the risk of sporadic prostate cancer. J Urol 179(5):2020–2024
Adam R, Spier I, Zhao B et al (2016) Exome sequencing identifies biallelic MSH3 germline mutations as a recessive subtype of colorectal adenomatous polyposis. Am J Hum Genet 99(2):337–351
Martelotto LG, Ng CK, de Filippo MR, Zhang Y, Piscuoglio S, Lim RS, Shen R, Norton L, Reis-Filho JS, Weigelt B (2014) Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Biol 15(10):484
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424
Association for Clinical Genetic Science (2016) Consensus statement on adoption of the American College of Medical Genetics and Genomics (ACMG) guidelines for sequence variant classification and interpretation [press release]. http://www.acgs.uk.com/media/1032817/acgs_consensus_statement_on_adoption_of_acmg_guidelines__1_.pdf. Accessed 10 Aug 2019
Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV, IARC Unclassified Genetic Variants Working Group (2008) Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 29(11):1282–1291
Vos J, Otten W, van Asperen C, Jansen A, Menko F, Tibben A (2008) The counsellees’ view of an unclassified variant in BRCA1/2: recall, interpretation, and impact on life. Psychooncology 17(8):822–830
Mersch J, Brown N, Pirzadeh-Miller S et al (2018) Prevalence of variant reclassification following hereditary cancer genetic testing prevalence of genetic variant reclassification after hereditary genes testing prevalence of genetic variant reclassification after hereditary genes testing. JAMA 320(12):1266–1274
Hoffman-Andrews L (2017) The known unknown: the challenges of genetic variants of uncertain significance in clinical practice. J Law Biosci 4(3):648–657
Murray ML, Cerrato F, Bennett RL, Jarvik GP (2011) Follow-up of carriers of BRCA1 and BRCA2 variants of unknown significance: variant reclassification and surgical decisions. Genet Med 13(12):998–1005
Otten E, Plantinga M, Birnie E, Verkerk MA, Lucassen AM, Ranchor AV, van Langen I (2015) Is there a duty to recontact in light of new genetic technologies? A systematic review of the literature. Genet Med 17(8):668–678
Seemanova E et al (2007) Cancer risk of heterozygotes with the NBN founder mutation. J Natl Cancer Inst 99(24):1875–1880
Funding
This study was supported and funded with thanks by the National Breast Cancer Research Institute (NBCRI), the Health Service Executive/Health Research Board National Specialist Academic Fellowship (NSAFP 2014/1), and the Monkstown Hospital Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: The originally published version of this article contained typesetting errors in Figure 2 and 3 legends. This has been corrected.
Rights and permissions
About this article
Cite this article
McVeigh, Ú.M., McVeigh, T.P., Curran, C. et al. Diagnostic yield of a custom-designed multi-gene cancer panel in Irish patients with breast cancer. Ir J Med Sci 189, 849–864 (2020). https://doi.org/10.1007/s11845-020-02174-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11845-020-02174-x