Abstract
In this study, 38 elite breeding pepper lines were genetically analyzed using SRAP markers and tested for resistance to PVY, TSWV, and PMMoV viruses using molecular markers. In the virus resistance tests, 1 line (37-H–D-6) from the Three-lobs population was found to be resistant to all 3 viruses tested. The 19 SRAP primer combinations used for genetic diversity yielded a total of 85 bands, 57 of which were polymorphic among pepper lines. While 2–8 bands per primer were obtained, the number of polymorphic bands ranged from 1 to 6. The average polymorphism rate of the primers was 66.44%. The PIC values ranged from 0.06 to 0.40 (with a mean of 0.18). In addition, the average gene diversity, effective allele number, and Shannon information index values of the primers were 0.21, 1.34, and 0.31, respectively. STRUCTURE analysis showed that the pepper lines were grouped into 4 clusters. PCoA and Q-matrix plots supported the cluster distribution. Some lines of the Sivri and Three-lobs pepper populations were observed as outliers in the plots. Kapia and Three-lobs were more similar to each other. This study showed that SRAP markers can be successfully used for genetic diversity of pepper breeding lines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Capsicum genus, which belongs to the Solanaceae family and is native to Central and South America with Bolivia as the proposed center of origin, is known to grow in temperate climates (van Zonneveld et al. 2015). Capsicum annuum, which is diploid and self-pollinating with a chromosome number of 2n = 24, is the most widely distributed and economically significant one among five domesticated Capsicum species (Moscone et al. 2007). It was introduced to Turkey in the sixteenth century through commercial interactions with Europeans, and eventually spread throughout Anatolia (Vural et al. 2000).
Genetic diversity is the basis of ecosystem biodiversity, including both intraspecific and interspecific diversity. There is high diversity among vegetable species based on consumer preferences. Many varieties are released annually for major vegetable species, with a focus on breeding for resistance to biotic (Pratap et al. 2020) and abiotic stresses (Shah et al. 2018), quality attributes, and productivity (do Rêgo et al. 2009). Although the variety development process takes many years, advances in biotechnology over the past 25–30 years have resulted in a reduction in breeding time. Genetic diversity studies using molecular methods allow breeders to rapidly characterize the genetic material of interest (Aktas et al. 2009; Bozokalfa et al. 2017; Yildiz et al. 2020), develop markers linked to desired traits (Li et al. 2011; Barka and Lee 2020), make selections using developed markers (Tanaka et al. 2014), and map species genomes (Paran et al. 2004). Restricted Fragment Length Polymorphism (RFLP) (Hyeon et al. 1997), Random Amplified Polymorphic DNA (RAPD) (Rêgo et al. 2011), Amplified Length Polymorphism (AFLP) (Lefebvre et al. 2001; Tam et al. 2005), microsatellite markers (Dhaliwal et al. 2013; Villela et al. 2014), Single-Nucleotide Polymorphism (SNP) (Du et al. 2019; Manivannan et al. 2021), and Sequence-Related Amplified Polymorphism (SRAP) (Xu et al. 2011; Qiu et al. 2021; Maulani et al. 2023) are the some molecular marker systems used in diversity of Capsicum germplasms. The use of advanced DNA-based methods in breeding allows breeders to easily overcome the challenges of the traditional breeding process and has reduced the time required to release new varieties.
SRAP technology is a marker system that preferentially amplifies open-reading frames (ORFs) and was first described in Brassica by Li and Quiros (2001). Two types of primers are used: forward primers (17 bp long with 14 nucleotides rich in C and G and 3 selective bases at the 3' end) and reverse primers (18 bp long with 15 nucleotides rich in A and T) (Ferriol et al. 2003). SRAP markers are useful for plant breeding programs and genetic studies due to their low cost and ease of production. They are highly polymorphic and can identify a large number of genetic variations within a population. This marker system has a range of applications in plant breeding, including identifying genetic differences between plants, evaluating the genetic diversity of breeding populations, and mapping genes (Kole et al. 2015; Bozokalfa et al. 2017; Jahangirzadeh et al. 2020). Genetic diversity is essential for the breeding and improvement of crop plants. Determining the relationship of local populations and breeding lines of the plant species in a germplasm, both morphologically and at the DNA level, is very important for the development of hybrid varieties.
Classical testing methods (Caranta and Palloix 1996; Romero et al. 2001) or molecular markers can be used to evaluate plant germplasm and breeding lines for disease and pest resistance. Molecular marker-assisted selection (MAS) allows breeders to identify individuals with specific resistance traits within a population (Moodley et al. 2019). In classical methods of testing resistance to viral diseases, disease resistance is assessed by the symptoms of the virus in the plant after a certain period of time following inoculation with the pathogen, depending on the inoculation method, the pathogen, and the environmental conditions. This method is both time-consuming and can affect the reliability of the results due to changes in symptoms depending on many factors. In addition, the inability to test for multiple diseases simultaneously in the same plant lengthens the breeding process and makes it difficult to develop lines and varieties resistant to multiple diseases. The development of MAS methods in breeding programs provides maximum information with minimum time and material input (Kumar et al. 2011). In particular, MAS enables the transfer of traits controlled by single genes (Collard and Mackill 2008; Jeong et al. 2015) and allows gene pyramiding in a variety to achieve the desired level of resistance (Özkaynak et al. 2014).
Vegetables have a very dynamic range of varieties, with many new varieties being released each year by breeding companies. In this situation, genetic resources and their evaluation are vital for breeders to adapt to this varietal dynamism. The aim of this study is to evaluate pepper breeding lines obtained from the Pepper Genetic Resources of the Black Sea Agricultural Research Institute (Samsun-Türkiye) for both genetic diversity and resistance to Potato Virus Y (PVY), Pepper Mild Mottle Virus (PMMoV), and Tomato Spotted Wilt Virus (TSWV) using molecular markers. By combining molecular marker analysis with resistance screening against these three economically important viral diseases, the study provides a comprehensive evaluation of breeding lines and makes the pepper gene pool with different fruit types and known genetic diversity available to vegetable breeders. This is particularly important for the breeding, as selecting parental lines that are genetically distant and have the desired traits increases the chances of success in developing superior hybrid varieties with a broad genetic base. Our approach demonstrates the potential to increase the efficiency of breeding efforts through the simultaneous evaluation of multiple traits, which facilitates the identification of promising parental lines for hybrid development and gene pyramiding strategies for the development of superior pepper varieties.
Materials and methods
Experiment was conducted using total 38 pepper lines and genotypes; 35 pepper breeding lines (at the S5 generation) obtained from Black Sea Agricultural Research Institute (Samsun-Türkiye) and 3 commercial pepper varieties (Table 1). The DNA isolation was performed according to Haymes (1996) miniprep protocol with minor modifications.
Screening of pepper lines for virus resistance
Pepper breeding lines were screened for PVY, TSWV, and PMMoV using CSO (Caranta et al. 1999), SCAC-568 (Moury et al. 2000), and AP-7/AP-8 (Matsunaga et al. 2003) markers, respectively. PCR reactions were performed using a total of 15 µl reaction mixture, consisting of 7.5 µl PCR master mix (Dream taq green master mix: Thermoscientific), 1 µl of forward and reverse primers (10 pmol each), 3 µl genomic DNA (15–20 ng), and 2.5 µl deionized water. The PCR protocol was as follows: pre-denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at the appropriate temperature for each primer for 30 s, and elongation at 72 °C for 60 s, and then post-elongation at 72 °C for 7 min. The PCR products of CSO and SCAC-568 primers were then restricted with AlwNI and XbaI restriction endonucleases, respectively, according to the manufacturer's (New England BioLabs Inc.) recommendations.
Characterization of pepper lines by SRAP markers
SRAP primers (Bioneer), selected from previous studies (Ferriol et al. 2003; Budak et al. 2004; Yildiz et al. 2011; Guo et al. 2014) and after our initial screening (14 forward and 10 reverse primers), were used to genetically evaluate the breeding lines. Totally, 144 primers pairs were initially screened and 19 of them were found to be polymorphic among pepper lines. PCR reactions were performed using total 15 µl of reaction mixture; 7.5 µl PCR master mix (Dream taq green master mix: Thermoscientific), 1 µl forward (ME) and 1 µl revers primers (EM) (10 pmol), 3 µl genomic DNA (15–20 ng), and 2.5 µl deionized water. PCR protocols were as follows; pre-denaturation at 94 °C for 2 min and followed for 5 cycles by denaturation at 94 °C 1 min, annealing at 35 °C for 1 min and elongation at 72 °C for 1 min, then denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min and elongation at 72 °C for 1 min by 35 cycles, and finally post-elongation at 72 °C for 5 min (Yildiz et al. 2011).
Electrophoresis
PCR products obtained from both the virus resistance assay and the SRAP evaluation were run on 3% of agarose gel (Fisher BioReagents) containing 1X TAE (Tris–Acetic Acid-EDTA) solution in SCIE-PLAS (Hu20) electrophoresis at 100 V and 300 mA currents for 4 h. After electrophoresis, the gel was stained with ethidium bromide (10 mg/ml) for 20 min, then washed with distilled water, and visualized under UV transilluminator (Syngene-Ingenius). Clear bands in the resulting gel image were scored as 1 (present) and 0 (absent), and then, a binary data matrix was generated for further data analysis.
Data analysis
The total number of fragments per primer and the number of polymorphic fragments per primer were scored and polymorphism rates (Pr) of the primers were calculated. The PopGene v.1.32 software (Yeh et al. 2000) was used to calculate genetic diversity parameters, such as the effective number of alleles (Ne) (Kimura and Crow 1964), gene diversity (H) (Nei 1973) and Shannon information index (I) (Lewontin 1972) for pepper breeding lines. The polymorphism information content (PIC) values and major allele frequency (Maf) values were obtained using PowerMarker v.3.25 software (Liu and Muse 2005). Clusters were created by UPGMA (unweighted pair group method with arithmetic mean) (Sneath 1979) and correlation matrix was generated using Dice module of NTSYSpc v.2.02 (Rohlf 2000) packaged program and then genetic relationship among pepper lines was also performed by using multivariate analysis. Principal Component Analysis (PCA) was realized using correlation matrix by NTSYSpc. Principal Coordinate Analysis (PCoA) was conducted in Past 3 statistical software. STRUCTURE analysis was performed for pepper lines using the Bayesian clustering algorithm in the STRUCTURE 2.3.4 software tool, as introduced by Pritchard et al. (2000) [length of burning period: 10,000 and number of Markov Chain Monte Carlo (MCMC) repeats after burning: 100,000 using 5 iterations].
Results
Screening of elite pepper lines for virus resistance
Thirty-five elite pepper breeding lines and three commercial varieties were screened for resistance to PVY, TSWV, and PMMoV using molecular markers. Resistance to different viruses has been observed in the Kapia (2 lines), Long-thin (1 line), Sivri (2 lines), and Three-lobs (3 lines) pepper populations (Table 1). Only one line 37-H–D-6 (Three-lobs) was found to be resistant to all three viruses. 68-H-12 (Kapia) and DM_3 were resistant to both PVY and TSWV. Among the other virus-resistant lines, 15-H-3–1 (Three-lobs) and 43H-7 (Kapia) were resistant only to PVY, while 68-H-1 (Three-lobs) and 13-H-1 (Sivri) were resistant only to TSWV. On the other hand, resistance to TMMoV was observed only in the breeding line 37-H–D-6 which was resistant to all 3 viruses.
Genetic characterization of pepper lines by SRAP markers
The 19 SRAP primers pairs generated a total 85 fragments and 57 of them (67.1%) were found to be polymorphic among pepper lines (Table 2). The total fragment numbers per locus varied between 2 (Me12/Em4, Me14/Em2, Me18/Em3) and 8 (Me1/Em2) (average 4.47 fragments per locus), while the polymorphic fragment number per locus was between 1 (Me12/Em4, Me14/Em11, Me18/Em3) and 6 (Me1/Em2, Me18/Em11) (average 3.00 fragments per locus). On the other hand, polymorphism rate (Pr) of the primers changed between 25% (Me14/Em11) and 100% (Me2/Em10, Me5/Em5, Me12/Em10, Me14/Em2). The major allele frequency values obtained from the SRAP primers were in the range of 0.64 (Me5/Em5, Me14/Em2) to 0.96 (Me10/Em4). Polymorphism information content (PIC) values of SRAP primers in pepper breeding lines were varied between 0.06 (Me10/Em4) and 0.40 (Me5/Em5) (average 0.18). Mean values of effective allele numbers (Ne), gene diversity (H), and Shannon information index (I) were found 1.34, 0.21, and 0.31, respectively. The primer combinations Me5/Em5 and Me14/Em2 gave the higher H (0.47 and 0.41) and I (0.67 and 0.59) values. These primer combinations effectively revealed the diversity among pepper inbred lines.
The principal component analysis (PCA) results revealed different patterns of genetic diversity among the pepper populations (Table 3). The eigenvalues associated with the first four principal components (PCs) were 33.09, 0.79, 0.67, and 0.51, respectively. These values indicate that the first PC explained most of the variance in the data set, accounting for 87.08% of the total variance. The subsequent components (PC2, PC3, and PC4) explain smaller proportions of the variance as the eigenvalues decrease. The cumulative variation explained by the first four PCs is 92.27%, indicating that these components together account for a large proportion of the genetic variation found in elite pepper breeding populations. In addition, the genetic diversity of our pepper breeding lines is clearly shown by the two-dimensional Principal Coordinate Analysis (PCoA) plot (Fig. 1). In the PCoA plot, it was clear that the Long-thin pepper lines and most of the Sivri peppers formed a distinct grouping, while the remaining lines had a larger spread across the plot. This grouping pattern indicates that the genetic variation among the Long-thin pepper lines is comparatively lower than that observed in the other pepper populations. In addition, several lines belonging to the Sivri pepper type and Three-lobs pepper populations were identified as outliers in the plot. These outliers suggest the presence of unique or divergent genetic profiles within these specific lines. This could be of particular interest for further investigation in the context of genetic improvement and breeding strategies. Indeed, 13-H-1 (Sivri) and 68-H-1 (Three-lobs) pepper lines were resistant to TSWV.
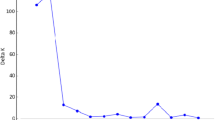
STRUCTURE analysis applied to SRAP marker data for genetic structure of elite pepper breeding lines revealed 4 major clusters. The Q-matrix obtained from STRUCTURE analysis represents the individual membership coefficients, indicating the proportion of an individual's genome that belongs to each of the inferred genetic clusters (Fig. 2). The average distance values among the pepper lines in each cluster were as follows: 0.1802 for cluster 1, 0.2555 for cluster 2, 0.1755 for cluster 3, and 0.0647 for cluster 4. This indicates that the similarity between the inbred lines in cluster 2 is lower than in the other clusters, while cluster 4 consists of the most similar pepper lines. The Mantel test value r for the UPGMA-based clustering analysis using the Dice similarity method was calculated to be 0.86. This value indicates that the breeding lines are distributed with a high degree of probability in the dendrogram. In the dendrogram constructed with the SRAP marker data, the majority of Long-thin pepper lines and Kapia pepper lines showed different clustering patterns, suggesting a closer genetic relationship within their respective clusters (Fig. 3). On the other hand, the Sivri and Three-lobs pepper lines showed a more scattered distribution among clusters. According to the dendrogram, Burkalem and Seyrek (Long-thin) were the closest pepper varieties, while 68-H-1 (Three-lobs) and 5-H-2 (Sivri) were the outlier elite pepper breeding lines, as shown in the PCoA plot (Fig. 1).
When conducting a genetic relationship analysis of 4 pepper populations, it was observed that Kapia and Three-lobs peppers exhibit a higher degree of genetic similarity (0.9439) between themselves compared to other pepper varieties (Table 4). The UPGMA-based clustering among the 4 pepper populations according to Nei’s genetic distance (Nei 1972) clearly showed this closer relation between Kapia and Three-lobs peppers (Fig. 4). Moreover, genetic similarity between Three-lobs peppers and Sivri peppers (0.9304) were nearly closer as between Kapia and Three-lobs peppers.
A dendrogram based on Nei’s (1972) genetic distance between 4 pepper populations
Discussion
One of the most important factors affecting pepper production worldwide is pests and diseases. In some regions where pepper is grown, disease management is a significant cost of production. Potato Virus Y (PVY) infects many solanaceous crops, including peppers (Ahirwar and Pachaya 2024). PVY can significantly reduce crop yield and quality, resulting in economic losses. Another important virus affecting peppers is Tomato Spotted Wilt Virus (TSWV), which is primarily transmitted by flower thrips (Wang et al. 2022). Because thrips are found inside pepper flowers with closed petals, chemical control is very difficult. In addition, although there are many viral pathogens for which pepper is a host, TSWV (Karavina and Gubba 2017) and Pepper Mild Mottle Virus (PMMoV) (Kumari et al. 2023) cause major problems, especially in greenhouse pepper production. Although cultural methods (Fajinmi and Odebode 2010; Karungi et al. 2013) are often used to control viral diseases in pepper, the most effective way to control viral diseases is genetic control (Sreenivas et al. 2020; Parisi et al. 2020), i.e., the development of virus-resistant varieties. In our study, the inbred line 37-H–D-6 possesses genes that confer resistance to PVY, TSWV, and PMMoV. In this respect, it is very important as a source for the development of virus-resistant hybrid varieties. In addition, several of our other inbred lines, including 68-H-12, DM-3, 15-H-3–1, 43H-7, 68-H-1, and 13-H-1, have the potential to be parents with resistance to various viruses.
It is very important to conserve plant genetic resources and to determine the genetic relationships between individuals and use them in breeding programs. Prediction of the genetic structure of genotypes by phenotypic observations is very difficult and this method is dependent on environmental conditions. The success of plant breeding activities worldwide has been increased by advances in molecular marker technologies (Nadeem et al. 2018), which have also significantly reduced the required time (Varshney et al. 2021). Therefore, molecular markers have become crucial for the rapid and reliable evaluation of plant genetic resources at the DNA level. Molecular markers work by highlighting differences (polymorphisms) in a nucleic sequence between different individuals. Various polymerase chain reaction (PCR) based molecular marker techniques are used in the genetic evaluation of plants. In the evaluation of pepper germplasm, several molecular marker systems, such as RAPD (Prasad et al. 2013; Tomlekova et al. 2023), ISSR (Inter Simple Sequence Repeats) (Alayachew et al. 2017; Srebcheva and Kostova 2023), SRAP (Du et al. 2010; Bozokalfa et al. 2017; Qiu et al. 2021), and microsatellite markers (Dhaliwal et al. 2014; Rabuma et al. 2020; Terefe et al. 2022), have been used. SRAP markers are a dominant marker system developed by Li and Quiros (2001). It has been successfully used to assess genetic diversity in many plant species, including pepper populations.
Du et al. (2010) compared different marker systems in the genetic characterization of pepper genotypes. In the study, it was reported that 29.6% of the bands obtained from 23 SRAP primers were polymorphic, with an average of 10 polymorphic bands per primer combination. The higher number of bands obtained here compared to the bands observed in our study is attributed to the researchers' ability to separate PCR products on polyacrylamide gels. However, our effective allele number per SRAP primer combination (average 1.34) was higher than the value reported in the mentioned study (1.14). Chen et al. (2012) reported that SRAP primer combinations were as successful as Simple Sequence Repeats (SSR) markers in their study on the genetic diversity of pepper genotypes. In the genetic evaluation of some pepper genotypes with resistance to P. capsici, it was observed that SRAP markers had a significantly higher mean number of polymorphic bands (5.00) compared to the results of our study (Göçmen and Abak 2019). However, the polymorphism rate of the primers was lower (60.77%) than our results. In another study using SRAP primer combinations for the molecular characterization of different pepper genotypes (Nas and İlbi 2022), it was reported that the number of polymorphic bands per primer combination ranged from 1 to 6. Zhong et al. (2021) reported that SSR makers revealed average PIC, MAF, NE, I, and H values were 0.29, 0.755, 1.705, 0.576, and 0.331 in C. frutescens, respectively. In a more recent study using SRAP markers in pepper (Maulani et al. 2023), the mean PIC, mean number of polymorphic bands, and mean PR were 0.338, 19.18, and 97.88%, respectively. Our results show that SRAP markers have a comparable success rate to SSR markers in the genetic analysis of pepper populations.
Principal component analysis (Jolliffe and Cadima 2016) is a powerful method for extracting information from genetic markers, because it can reduce multivariate genetic information into a small number of synthetic variables (Abdi and Williams 2010). The results of PCA can lead to a better understanding of genetic diversity and structure of the pepper populations using that SRAP primers. Nas and İlbi (2022) reported that the first component in the PCA applied to the data obtained with SRAP markers explained 69.91% and 70.49% of the variation for banana pepper and bell pepper lines, respectively. In another study, the first PC axis explained 65% of the total variance as determined by PCA results obtained from SRAP marker data in the diversity of Turkish pepper genotypes (Bozokalfa et al. 2017). Our PCA results revealed by SRAP marker data were higher informative than those results. This could be due to several factors, including greater genetic coverage and improved discriminatory power of the SRAP primer combinations we used. STRUCTURE analysis applied to SRAP marker data for genetic structure of elite pepper breeding lines revealed 4 major clusters. In a study conducted with SRAP markers to investigate the genetic differences of oil palm populations (Zhou et al. 2021), STRUCTURE analysis showed the best K means for oil palm populations was 3. On the other hand, in a genetic screening study with SSR markers in pepper genotypes (Guo et al. 2023), STRUCTURE analysis results showed that the best delta K value was 2. Based on the dendrogram, Burkalem and Seyrek (Long-thin pepper) were the closest pepper varieties, while 68-H-1 (Three-lobs) and 5-H-2 (Sivri pepper) were the most divergent elite pepper breeding lines. Our results are in agreement with the results of previous studies that have used SRAP markers to evaluate the genetic diversity of pepper accessions. Xu et al. (2011) found satisfying levels of genetic diversity among 72 pepper accessions using SRAP markers, and these accessions were further divided into 8 groups. Although the SRAP markers are dominant, they have been reported to provide reliable results that are comparable to those obtained with co-dominant SSR markers in distinguishing pepper populations (Chen et al. 2012). The UPGMA-based clustering among the 4 pepper populations (types) clearly showed this closer relation between Kapia and Three-lobs peppers. These findings confirm the results of PCoA and STRUCTURE analysis.
Conclusions
Regarding the resistance of pepper lines to PVY, TSWV, and PMMoV, it was observed that at least one line in each pepper population was resistant to at least one virus. There was one breeding line (37-H–D-6) in the Three-lobs pepper population that was resistant to all three viruses. This breeding line shows a very good parental potential for the development of F1 hybrid varieties resistant to multiple virus diseases. The genetic diversity of pepper genotypes is an important factor for breeding programs. SRAP markers are useful tools for assessing the genetic diversity and relationships of pepper genotypes, as they are reproducible and polymorphic. In this study, the high explanatory value of the first axis of the PCA suggests that the SRAP primers are particularly informative and have a strong screening capacity in the diversity of different pepper breeding lines. Therefore, the genetic diversity and relationships of four pepper types (Long-thin, Kapia, Sivri, and Three-lobs) were successfully investigated using SRAP markers. The results showed that Long-thin peppers and Kapia peppers had distinct clustering patterns, suggesting a closer genetic relationship within their respective groups. Sivri and Three-lobs peppers showed a more scattered distribution across several clusters, indicating a higher genetic variation among these types. Some of these were identified as outliers (68-H-1 and 5-H-2). These outliers indicate the presence of unique or divergent genetic profiles within these specific lines, which could be of particular interest for further investigation in the context of genetic improvement and breeding strategies. The genetic similarity analysis showed that Kapia and Three-lobs had the highest genetic similarity among the four populations.
Data availability
All relevant data are included within the manuscript. Any additional information required to replicate the findings of this study is available upon request from the corresponding author.
References
Abdi H, Williams LJ (2010) Principal component analysis. Wiley Interdiscip Rev Comput Stat 2:433–459. https://doi.org/10.1002/wics.101
Ahirwar NK, Pachaya JS (2024) Effects of climate change on the spread and severity of Potato Virus Y: an in-depth examination. Asian J Microbiol Biotechnol 9:39–59. https://doi.org/10.56557/ajmab/2024/v9i18605
Aktas H, Abak K, Sensoy S (2009) Genetic diversity in some Turkish pepper (Capsicum annuum L.) genotypes revealed by AFLP analyses. African Journal of Biotechnology 8(18): 4378–4386
Alayachew S, Atnafu D, Gedefa S (2017) Genetic diversity study of Ethiopian hot pepper cultivars (Capsicum spp.) using Inter Simple Sequence Repeat (ISSR) marker. J Plant Mol Breed 5(2):27–37. https://doi.org/10.22058/jpmb.2019.91684.1169
Barka GD, Lee J (2020) Molecular marker development and gene cloning for diverse disease resistance in pepper (Capsicum annuum L.): current status and prospects. Plant Breed Biotechnol 8(2):89–113. https://doi.org/10.9787/PBB.2020.8.2.89
Bozokalfa MK, Aşcıoğlu TK, Eşiyok D (2017) Biber genotiplerinin genetik çeşitliliklerinin SRAP markörleri kullanılarak belirlenmesi. Anadolu Tarım Bilimleri Dergisi 32(3):321–329. https://doi.org/10.7161/omuanajas.284511
Budak HR, Shearman C, Parmaksiz I, Gaussoin RE, Riordan TP, Dweikat I (2004) Molecular characterization of buffalograss germplasm using sequence-related amplified polymorphism markers. Theor Appl Genet 108:328–334. https://doi.org/10.1007/s00122-003-1428-4
Caranta C, Palloix A (1996) Both common and specific genetic factors are involved in polygenic resistance of pepper to several potyviruses. Theor Appl Genet 92(1):15–20. https://doi.org/10.1007/BF00222946
Caranta C, Thabuis A, Palloix A (1999) Development of a CAPS marker for the Pvr4 locus: a tool for pyramiding potyvirus resistance genes in pepper. Genome 42(6):1111–1116. https://doi.org/10.1139/g99-069
Chen XJ, Zong ZKH, HX, Fang R, (2012) Genetic diversity of Capsicum frutescens in China as revealed by SRAP and SSR markers. Acta Botan Boreali-Occiden Sin 32(11):2201–2205
Collard BCY, Mackill DJ (2008) Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363:557–572. https://doi.org/10.1098/rstb.2007.2170
Dhaliwal MS, Jindal SK, Gaikwad AK, Singh K (2013) Genetic diversity analysis and DNA fingerprinting of elite chilli pepper lines using SSR markers. Int J Veget Sci 19(3):207–216. https://doi.org/10.1080/19315260.2012.707762
Dhaliwal MS, Yadav A, Jindal SK (2014) Molecular characterization and diversity analysis in chilli pepper using simple sequence repeats (SSR) markers. Afr J Biotech 13(31):3137–3143. https://doi.org/10.5897/AJB2014.13695
do Rêgo ER, do Rêgo MM, Finger FL, Cruz CD, Casali VWD, (2009) A diallel study of yield components and fruit quality in chilli pepper (Capsicum baccatum). Euphytica 168:275–287. https://doi.org/10.1007/s10681-009-9947-y
Du X, Wang D, Gong Z (2010) Comparison of RSAP, SRAP and SSR markers for genetic analysis in hot pepper. Indian J Horticulture 64(4b):505–512
Du H, Yang J, Chen B, Zhang X, Zhang J, Yang K, Wen C (2019) Target sequencing reveals genetic diversity, population structure, core-SNP markers, and fruit shape-associated loci in pepper varieties. BMC Plant Biol 19(1):1–16. https://doi.org/10.1186/s12870-019-2122-2
Fajinmi AA, Odebode CA (2010) Evaluation of maize/pepper intercropping model in the management of pepper veinal mottle virus, genus Potyvirus, family Potyviridae on cultivated pepper (Capsicum annuum L.) in Nigeria. Arch Phytopathol Plant Prot 43(15):1524–1533. https://doi.org/10.1080/03235400802583677
Ferriol M, Picó B, Nuez F (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genet 107:271–282. https://doi.org/10.1007/s00122-003-1242-z
Göçmen M, Abak K (2019) Phytophthora capsici’ye dayanıklı bazı biber genotiplerinin SRAP ve SSR belirteçlerle genetik farklılıklarının belirlenmesi. Derim 36(2):124–134. https://doi.org/10.16882/derim.2019.557877
Guo Y, Lin H, Liu Z, Zhao Y, Guo X, Li K (2014) SSR and SRAP marker-based linkage map of Vitis vinifera L. Biotechnol Biotechnol Equip 28(2):221–229. https://doi.org/10.1080/13102818.2014.907996
Guo G, Pan B, Yi X, Khan A, Zhu X, Ge W, Liu J, Diao W, Wang S (2023) Genetic diversity between local landraces and current breeding lines of pepper in China. Sci Rep 13(1):4058. https://doi.org/10.1038/s41598-023-29716-4
Haymes KM (1996) Mini-prep method suitable for a plant breeding program. Plant Mol Biol Report 14(3):280–284. https://doi.org/10.1007/BF02671664
Hyeon NS, Woong YJ, Cheorl KB, Byung-Dong K (1997) Selection of parental lines for hot pepper mapping population using RFLP and AFLP analyses. Hortic Environ Biotechnol 38(6):693–697
Jahangirzadeh S, Azadi Gonbad R, Falakro K (2020) Identification of genetic diversity and relationships of some Iranian tea genotypes using SRAP markers. J Horticult Postharv Res 3(1):25–34. https://doi.org/10.22077/jhpr.2019.2582.1067
Jeong HS, Jang S, Han K, Kwon JK, Kang BC (2015) Marker-assisted backcross breeding for development of pepper varieties (Capsicum annuum) containing capsinoids. Mol Breeding 35(12):1–10. https://doi.org/10.1007/s11032-015-0417-z
Jolliffe IT, Cadima J (2016) Principal component analysis: A review and recent developments. Philos Trans R Soc A Math Phys Eng Sci 374:20150202. https://doi.org/10.1098/rsta.2015.0202
Karavina C, Gubba A (2017) Detection and characterization of Tomato spotted wilt virus infecting field and greenhouse-grown crops in Zimbabwe. Eur J Plant Pathol 149:933–944. https://doi.org/10.1007/s10658-017-1243-4
Karungi J, Obua T, Kyamanywa S, Mortensen CN, Erbaugh M (2013) Seedling protection and field practices for management of insect vectors and viral diseases of hot pepper (Capsicum chinense Jacq.) in Uganda. Int J Pest Manag 59(2):103–110. https://doi.org/10.1080/09670874.2013.772260
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49(4):725–738. https://doi.org/10.1093/genetics/49.4.725
Kole PR, Sharma MK, Kumar S, Kumar A, Singh S, Sengar RS (2015) Assessment of genetic diversity in Indian cultivated pea (Pisum Sativum L.) by using SRAP markers. Int J Plant Res 28(3):84–91. https://doi.org/10.5958/2229-4473.2015.00070.1
Kumar J, Choudhary AK, Solanki RK, Pratap A (2011) Towards marker-assisted selection in pulses: A review. Plant Breed 130(3):297–313. https://doi.org/10.1111/j.1439-0523.2011.01851.x
Kumari N, Sharma V, Patel P, Sharma PN (2023) Pepper mild mottle virus: a formidable foe of capsicum production—a review. Front Virol 3:1208853. https://doi.org/10.3389/fviro.2023.1208853
Lefebvre V, Goffinet B, Chauvet JC, Caromel B, Signoret P, Brand R, Palloix A (2001) Evaluation of genetic distances between pepper inbred lines for cultivar protection purposes: comparison of AFLP, RAPD and phenotypic data. Theor Appl Genet 102(5):741–750. https://doi.org/10.1007/s001220051705
Lewontin RC (1972) Testing the theory of natural selection. Nature 236(5343):181–182. https://doi.org/10.1038/236181a0
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461. https://doi.org/10.1007/s001220100570
Li X, Yang H, Buirchell B, Yan G (2011) Development of a DNA marker tightly linked to low-alkaloid gene iucundus in narrow-leafed lupin (Lupinus angustifolius L.) for marker-assisted selection. Crop Pasture Sci 62(3):218–224. https://doi.org/10.1071/CP10352
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinform 21:2128–2129. https://doi.org/10.1093/bioinformatics/bti282
Manivannan A, Choi S, Jun TH, Yang EY, Kim JH, Lee ES, Lee HE, Kim DS, Ahn YK (2021) Genotyping by sequencing-based discovery of SNP markers and construction of linkage map from F5 population of pepper with contrasting powdery mildew resistance trait. Biomed Res Int 2021:6673010. https://doi.org/10.1155/2021/6673010
Matsunaga H, Saito T, Hirai M, Nunome T, Yoshida T (2003) DNA markers linked to Pepper mild mottle virus (PMMoV) resistant locus (L4) in Capsicum. J Jap Soc Hort Sci 72(3):218–220. https://doi.org/10.2503/jjshs.72.218
Maulani R, Murti RH, Purwantoro A (2023) Molecular diversity in populations of chili (Capsicum annuum L.). Sabrao J Breed Genet 55(1):15–24. https://doi.org/10.54910/sabrao2023.55.1.2
Moodley V, Naidoo R, Gubba A, Mafongoya PL (2019) Development of Potato virus Y (PVY) resistant pepper (Capsicum annuum L.) lines using marker-assisted selection (MAS). Physiol Mol Plant Pathol 105:96–101. https://doi.org/10.1016/j.pmpp.2018.12.002
Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sanchez García Y, Jarret R, Daviña JR, Ducasse DA, Barboza GE, Ehrendorfer F (2007) The evolution of chili peppers (Capsicum-Solanaceae): a cytogenetic perspective. Acta Hort 745:137–170. https://doi.org/10.17660/ActaHortic.2007.745.5
Moury B, Pflieger S, Blattes A, Lefebvre V, Palloix A (2000) A CAPS marker to assist selection of tomato spotted wilt virus (TSWV) resistance in pepper. Genome 43(1):137–142. https://doi.org/10.1139/g99-098
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Baloch FS (2018) DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32(2):261–285. https://doi.org/10.1080/13102818.2017.1400401
Nas Y, İlbi H (2022) Determination of genetic diversity in banana and bell pepper lines using molecular markers. Uluslararası Tarım Ve Yaban Hayatı Bilimleri Dergisi 8(2):234–244. https://doi.org/10.24180/ijaws.1098482
Nei M (1972) Genetic distance between populations. Am Nat 106(949):283–292. https://doi.org/10.1086/282771
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70(12):3321–3323. https://doi.org/10.1073/pnas.70.12.3321
Özkaynak E, Devran Z, Kahveci E, Doganlar S, Basköylü B, Dogan F, İsleyen M, Yüksel A, Yüksel M (2014) Pyramiding multiple genes for resistance to PVY, TSWV and PMMoV in pepper using molecular markers. Eur J Hortic Sci 79(4):233–239
Paran I, van der Voort JR, Lefebvre V, Jahn M, Landry L, van Schriek M, Tanyolac B, Caranta C, Ben Chaim A, Livingstone K, Palloix A, Peleman J (2004) An integrated genetic linkage map of pepper (Capsicum spp.). Mol Breeding 13:251–261. https://doi.org/10.1023/B:MOLB.0000022526.30914.31
Parisi M, Alioto D, Tripodi P (2020) Overview of biotic stresses in pepper (Capsicum spp.): Sources of genetic resistance, molecular breeding and genomics. Int J Mol Sci 21(7):2587. https://doi.org/10.3390/ijms21072587
Prasad B, Khan RG, Radha T, Ravi C, Venkataiah P, Subhash K, Reuben TC (2013) DNA profiling of commercial chilli pepper (Capsicum annuum L.) varieties using random amplified polymorphic DNA (RAPD) markers. Afr J Biotechnol 12(30):4730–4735. https://doi.org/10.5897/AJB2012.3017
Pratap A, Douglas C, Prajapati U, Kumari G, RasheedWar A, Tomar R, Pandey AK, Dubey S (2020) Breeding Progress and Future Challenges: Biotic Stresses. In: Nair R, Schafleitner R, Lee SH (eds) The Mungbean Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-030-20008-4_5
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959. https://doi.org/10.1093/genetics/155.2.945
Qiu YH, Lin ST, Zeng SG, Luo Y, Zhu BT, Li YQ (2021) Phenotypic and SRAP genetic diversity analysis of 41 yellow pepper germplasm resources. J South Agric 52(8):2165–2173. https://doi.org/10.3969/j.issn.2095-1191.2021.08.015
Rabuma T, Gupta OP, Chhokar V (2020) Phenotypic characterization of chili pepper (Capsicum annuum L.) under Phytophthora capsici infection and analysis of genetic diversity among identified resistance accessions using SSR markers. Physiol Mol Plant Pathol 112:101539. https://doi.org/10.1016/j.pmpp.2020.101539
Rêgo ER, Rego MM, Farias-Filho LP (2011) Genetic diversity in pepper (Capsicum spp.) by RAPD marker. Acta Hort 918:341–347. https://doi.org/10.17660/ActaHortic.2011.918.44
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.1. Exeter Software, New York
Romero A, Blanco-Urgoiti B, Soto MJ, Fereres A, Ponz F (2001) Characterization of typical pepper-isolates of PVY reveals multiple pathotypes within a single genetic strain. Virus Res 79(1–2):71–80. https://doi.org/10.1016/S0168-1702(01)00300-8
Shah LR, Sharma A, Nabi J, Rathore JP (2018) Breeding approaches for abiotic stress management in vegetable crops. J Pharmacogn Phytochem 7(3):1023–1028
Sneath PHA (1979) BASIC program for a significance test for clusters in UPGMA dendrograms obtained from squared Euclidean distances. Comput Geosci 5(1):127–137. https://doi.org/10.1016/0098-3004(79)90020-7
Srebcheva TA, Kostova ML (2023) Investigation on genetic diversity of pepper (Capsicum spp.) parents and interspecific hybrids using ISSR markers. Genetika 55:883–893. https://doi.org/10.2298/GENSR2303883S
Sreenivas M, Bhattacharjee T, Sharangi AB, Maurya PK, Banerjee S, Chatterjee S, Maji A, Kumar Mandal A, Chakraborty I, Chattopadhyay A (2020) Breeding chili pepper for simultaneous improvement in dry fruit yield, fruit quality and leaf curl virus disease tolerance. Int J Veg Sci 26(5):457–486. https://doi.org/10.1080/19315260.2019.1648351
Tam SM, Mhiri C, Vogelaar A, Kerkveld M, Pearce SR, Grandbastien MA (2005) Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theor Appl Genet 110(5):819–831. https://doi.org/10.1007/s00122-004-1837-z
Tanaka Y, Yoneda H, Hosokawa M, Miwa T, Yazawa S (2014) Application of marker-assisted selection in breeding of a new fresh pepper cultivar (Capsicum annuum) containing capsinoids, low-pungent capsaicinoid analogs. Sci Hortic 165:242–245. https://doi.org/10.1016/j.scienta.2013.10.025
Terefe M, Alemu SK, Olani G, Debebe A, Aklilu S, Berhanu B (2022) Genetic diversity in pepper (Capsicum annum L.) germplasms using SSR markers. Afr J Plant Sci 16(7):174–185
Tomlekova N, Spasova-Apostolova V, Panayotov N, Panchev I, Sarsu F, Wani MR, Seibt KM (2023) Applicability of ISAP and RAPD techniques for Capsicum collection genotyping. In: Advanced crop improvement, vol 2: Case studies of economically important crops. Springer, Cham, pp 77–114. https://doi.org/10.1007/978-3-031-26669-0_3
van Zonneveld M, Ramirez M, Williams DE, Petz M, Meckelmann S, Avila T, Bejarano C, Ríos L, Peña K, Jäger M, Libreros D, Amaya K, Scheldeman X (2015) Screening genetic resources of Capsicum peppers in their primary center of diversity in Bolivia and Peru. PLoS ONE 10(9):e0134663. https://doi.org/10.1371/journal.pone.0134663
Varshney RK, Bohra A, Yu J, Graner A (2021) Designing future crops: genomics-assisted breeding comes of age. Trends Plant Sci 26:631–649. https://doi.org/10.1016/j.tplants.2021.03.010
Villela JCB, Barbieri RL, Castro CM, Neitzke RS, Vasconcelos CS, Carbonari T, Mistura CC, Priori D (2014) Caracterização molecular de variedades crioulas de pimentas (Capsicum baccatum) com marcadores microssatélites. Hortic Bras 32:131–137. https://doi.org/10.1590/S0102-0536201400020002
Vural H, Esiyok D, Duman I (2000) Kültür Sebzeleri (Sebze Yetiştirme). Ege Üniversitesi Basımevi, Bornova, İzmir:440 s.
Wang H, Wu X, Huang X, Wei S, Lu Z, Ye J (2022) Seed transmission of tomato spotted wilt orthotospovirus in peppers. Viruses 14:1873. https://doi.org/10.3390/v14091873
Xu X, Liu Z, Lin X, Mou S, Guan D, He S (2011) Genetic diversity and relationship analysis of pepper germplasm resources based on phenotype traits and SRAP molecular markers. J Fujian Agric For Univ 40(1):48–53
Yeh FC, Yang RC, Boyle TJ, Ye Z, Xiyan JM, Yang R (2000) POPGENE 32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton, Microsoft Windows-based freeware for population genetic analysis
Yildiz M, Ekbic E, Keles D, Sensoy S, Abak K (2011) Use of ISSR, SRAP, and RAPD markers to assess genetic diversity in Turkish melons. Sci Hortic 130(1):349–353. https://doi.org/10.1016/j.scienta.2011.06.048
Yildiz M, Kocak M, Nadeem MA, Cavagnaro P, Barboza K (2020) Genetic diversity analysis in the Turkish pepper germplasm using iPBS retrotransposon-based markers. Turk J Agric for 44(1):1–14. https://doi.org/10.3906/tar-1902-10
Zhong Y, Yuang C, Meiying R, Qingjing Y, Rongqing W, Zhuping Y, Guozhi Z, Jia L, Jiahong Y, Hongjian W (2021) High-throughput SSR marker development and the analysis of genetic diversity in Capsicum frutescens. Horticulturae 7(7):187. https://doi.org/10.3390/horticulturae7070187
Zhou L, Yarra R, Cao H, Zhao Z (2021) Sequence-related amplified polymorphism (SRAP) markers based genetic diversity and population structure analysis of oil palm (Elaeis guineensis Jacq.). Trop Plant Biol 14:63–71. https://doi.org/10.1007/s12042-020-09273-0
Acknowledgements
This study was supported financially by Ordu University, project code: BY-1733.
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ekbiç, E., Okay, C.Ö. Assessment of elite pepper breeding lines using molecular markers. Plant Biotechnol Rep (2024). https://doi.org/10.1007/s11816-024-00911-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11816-024-00911-7