Abstract
Evolvability is best addressed from a multi-level, macroevolutionary perspective through a comparative approach that tests for among-clade differences in phenotypic diversification in response to an opportunity, such as encountered after a mass extinction, entering a new adaptive zone, or entering a new geographic area. Analyzing the dynamics of clades under similar environmental conditions can (partially) factor out shared external drivers to recognize intrinsic differences in evolvability, aiming for a macroevolutionary analog of a common-garden experiment. Analyses will be most powerful when integrating neontological and paleontological data: determining differences among extant populations that can be hypothesized to generate large-scale, long-term contrasts in evolvability among clades; or observing large-scale differences among clade histories that can by hypothesized to reflect contrasts in genetics and development observed directly in extant populations. However, many comparative analyses can be informative on their own, as explored in this overview. Differences in clade-level evolvability can be visualized in diversity-disparity plots, which can quantify positive and negative departures of phenotypic productivity from stochastic expectations scaled to taxonomic diversification. Factors that evidently can promote evolvability include modularity—when selection aligns with modular structure or with morphological integration patterns; pronounced ontogenetic changes in morphology, as in allometry or multiphase life cycles; genome size; and a variety of evolutionary novelties, which can also be evaluated using macroevolutionary lags between the acquisition of a trait and phenotypic diversification, and dead-clade-walking patterns that may signal a loss of evolvability when extrinsic factors can be excluded. High speciation rates may indirectly foster phenotypic evolvability, and vice versa. Mechanisms are controversial, but clade evolvability may be higher in the Cambrian, and possibly early in the history of clades at other times; in the tropics; and, for marine organisms, in shallow-water disturbed habitats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Not all traits, populations, species, or clades have been equally labile or productive over their evolutionary lifetimes. A fundamental challenge lies in understanding the basis of those contrasts, particular in distinguishing the role of intrinsic factors at various levels, from the configuration of gene-regulatory networks in an organism to the geographic extent of a clade, and extrinsic factors, from local competition to global climatic upheavals, in determining such differences. (See Jablonski, 2017a, 2017b for a general discussion of intrinsic and extrinsic factors in macroevolution, i.e., evolution above the species level.) One potential intrinsic factor is evolvability, an evolutionary property that has become a major focus of discussion and analysis, primarily from developmental and quantitative genetic viewpoints (Hansen et al., 2022).
Evolvability has been defined in many ways (see Brown, 2014; Nuño de la Rosa, 2017; Houle & Pélabon, 2022), but when couched in general terms—the disposition or propensity to evolve, often referring specifically to adaptive evolution—it can reside at any level within the biological hierarchy. In the macroevolutionary perspective adopted here, the focus will be on species and clades. Addressing evolvability at this level, requires analyses that test (a) whether species and clades vary in their intrinsic evolvability, and if so, (b) what determines that variation, and (c) whether those intrinsic differences are stable over a clade’s history. Viewed from the other direction, we need to determine whether the genetic and developmental mechanisms thought to promote evolvability in the short term have predictable long-term, large-scale evolutionary consequences. Given among-clade variation in evolvability, we can even ask, in the canonical terms of evolution by natural selection, whether that variation imposes differential survival and reproduction of evolutionary units, and whether that variation is heritable at the relevant level of organization. If so, then selection among clades for evolvability is feasible (see, for example, Gerhart & Kirschner, 1997; Draghi & Wagner, 2008: Hansen, 2011: p. 369; Lehman & Stanley, 2013). This is a challenging agenda, because inferences at the requisite scale and hierarchical level almost always rely on indirect evidence. Here I outline macroevolutionary approaches to evolvability, first regarding intrinsic traits that may enhance or reduce evolvability among clades, with some discussion of traits that might themselves be more evolvable, and then regarding variation in evolvability across time and space. This paper cannot provide definitive answers, but aims to present an operational macroevolutionary approach, and to organize questions and potential examples to stimulate further theoretical and empirical research.
Operationalizing Evolvability in Macroevolution
The term evolvability might apply to any macroevolutionary currency, such as taxonomic diversity, functional variety, or morphological disparity, indeed a long-standing question has been the degree of covariation among those currencies in different situations (Folk et al., 2019; Jablonski, 2017a, 2017b; Martin & Richards, 2019; Shi et al., 2021). I propose to confine evolvability in macroevolution to phenotypes, with the hypothesis that evolvability is manifested in the behavior of traits and clades in a quantitative morphospace or functional space. An enormous literature exists on factors that promote or damp speciation and taxonomic diversification, but the propensity to achieve reproductive isolation, or to accrue taxonomic richness, probably involves a very different set of organismal and species-level attributes from those promoting the evolvability of form or function (see for example Jablonski, 2008a, 2017b; Rundell & Price, 2009; Harvey et al., 2019). Thus, treating differential taxonomic rates or patterns as another aspect of evolvability probably does not gain much.
For macroevolutionary purposes, we can operationalize evolvability as the differential (phenotypic) ability to take advantage of, or respond to, opportunity. This comparative approach is broadly analogous to the measurement of evolvability in terms of differential responses of traits to a unit strength of directional selection (Hansen & Pélabon, 2021). Both intrinsic and extrinsic factors can create the opportunities—the acquisition of a novel structure, developmental pathway, or mode of life; entry into a novel ecosystem by surviving a mass extinction, invading a new landmass, or encountering newly evolved or introduced resources—and the analysis entails comparison of how clades performed in response (for useful discussions of evolutionary opportunity, see Losos, 2010; Gillespie et al., 2020). The difficulty for macroevolutionary analysis, of course, is that no two convergent evolutionary novelties are truly identical, and no two clades are likely to experience an environment in identical ways. Contingency and context matter, especially at these temporal and spatial scales. But because we can set prior expectations for the consequences of at least some confounding factors, we can frame hypotheses incorporating them, approaching macroevolutionary.
This phenotypic approach, predicated on net phenotypic shifts or gains of disparity in morphology or function, also differs from a view of evolvability as the ability of a species or clade to realize variation in any direction from a starting phenotype (i.e. minimal developmental bias; Uller et al., 2018). The “bias” approach would allow clades to be evaluated in isolation and perhaps may be useful over short timescales, but is often insufficient for macroevolutionary purposes. Many clades traced through multivariate morphospaces (“phylomorphospaces”) undergo much movement in morphospace with little net expansion or shift compared to related clades; such contrasts are seen, for example, in sister clades in anostomoid fishes (Sidlauskas, 2008) and in the major branches of post-Paleozoic echinoids (Hopkins & Smith, 2015; Fig. 1). Thus, when assessing evolvability using clades’ behavior in morphospace disparity metrics based on density of morphospace occupation, such as pairwise distances, will be less informative than those measuring the spread, such as convex-hull volume or mean distance from the centroid (see Guillerme, Cooper, et al., 2020; Guillerme, Puttick, et al., 2020).
Differences in apparent evolvability in the major sea-urchin clades, portrayed in a phylomorphospace based on principal coordinates analysis of a character matrix. Modified after Hopkins and Smith (2015), used by permission (Color figure online)
Similarly, frequent changes in discrete characters, even if apparently isotropic around a given starting point, need not yield extensive net change when homoplasy is common across the phylogeny. Then, character-state exhaustion in the descriptive sense—conceivably with an underlying genetic basis in some cases—can be seen in clades showing many state changes but low macroevolutionary evolvability in terms of capture of new states (as recognized decades ago, see Foote, 1997; Wagner, 2000; and see Oyston et al., 2015, who find that such exhaustion cannot explain temporal changes in clades’ behavior in morphospace). This is one reason for heterogenous results on the correlation between (morpho)speciation rates and overall phenotypic evolution: much total change can occur while repeatedly traversing a limited range of morphologies; another possible reason is that traits serving as the raw material for phylogenetic analyses may not be the ones that govern lineage splitting and persistence (see Crouch, 2020). The larger question remains: whether or how often among-clade differences in apparent evolvability can be understood, and predicted, in terms of intrinsic differences rather than simply reflecting the operation of extrinsic pressures. Of course, the intrinsic-extrinsic distinction is not clearly demarcated and both factors often operate in concert to some degree; but the above-mentioned macroevolutionary “common-garden” approach can help to identify intrinsic among-clade differences, such as those governing differential responses to opportunities opened to clades following extinction events.
The two major arenas for macroevolutionary analysis—the fossil record and comparative data on extant taxa—are essentially historical or retrospective, each with different strengths and weaknesses; they are most powerful when applied in concert, although integrating them is difficult (e.g. Jablonski, 2017b; Mitchell et al., 2019; Quental & Marshall, 2010). Neontological approaches (mostly) begin with genetic or developmental data thought to indicate evolvability and attempt to recognize how they have shaped the large-scale dynamics of the clade leading to the present day; paleontological analyses (mostly) begin with the phenotypic dynamics and attempt to exclude confounding factors to recognize differences in intrinsic evolvability among clades. Despite their imperfections, both approaches at the very least reveal intriguing phenomena worthy of deeper investigation. In either domain the first step is to frame comparative analyses, potentially identifying the role of intrinsic biological properties relative to the myriad extrinsic factors that can drive differences in evolutionary tempo and mode among clades in time and space. (See for example the sea urchin example below, Fig. 1, with contrasting macroevolutionary patterns in the major subclades and a potential developmental mechanism for the persistent differences in evolvabiilty among them).
Observations on Extant Organisms
As noted, one approach measures attributes in extant populations that might impose or reflect differing degrees of evolvability of traits or clades, and then tests predictions retrospectively, i.e. by analyzing macroevolutionary outcomes or estimated dynamics of those traits or clades (e.g. Goswami & Polly, 2010 on primates vs carnivores, with important later work incorporating extensive fossil data; Haber, 2016 on ruminants; Houle et al., 2017 on Drosophila). Such analyses require some strong or poorly understood assumptions. For example, the stability of G-matrices describing genetic (co)variances tied to phenotypes is uncertain at these scales, particularly in light of the complex, nonlinear relationships between genotype and phenotype as quantified in G-matrices and P-matrices (Milocco & Salazar-Ciudad, 2022). Thus, the utility of extrapolating from present-day data (Hansen & Pélabon, 2021), and their roles in determining properties such as modularity or the isotropy of accessible phenotypes around a given starting point, is debated, with a variety of empirical outcomes. Further analyses in a multispecies phylogenetic framework would be valuable (see Love et al., 2021; and Saltzberg et al., 2022, relatively narrow in its phenotypic and phylogenetic coverage but encouraging in not finding a correlation between G-matrix divergence and time since species splits). Urgently needed for such cross-scale applications is an array of genetic and developmental model systems that have robust fossil records. Most existing models are both highly derived and paleontologically scant, and thus uninformative for many types of macroevolutionary analysis. Progress is certainly being made, but model systems involving a decapod or ostracode crustacean instead of Drosophila, an irregular echinoid instead of Strongylocentrotus, or a bivalve instead of the squid Euprymna, would be an enormous step toward integrating the two great branches of historical biology.
A further assumption required to extrapolate from modern organisms to macroevolutionary scales is that taxonomic or morphological dynamics can be robustly derived from the topology of large molecular phylogenies. Here too some progress has been made, but separating speciation and extinction rates from net diversification—potentially important for testing hypotheses of cause and effect in morphospace occupation (as in Huang et al., 2015)—remains challenging (e.g. Louca & Pennell, 2020; Love et al., 2021), as does the problem of inferring ancestral character states from extant taxa alone (Betancur-R et al., 2015; Marshall, 2017; Slater et al., 2012); and as Mangiardino Koch (2021) notes more generally (citing twelve studies), evolutionary modeling is demonstrably improved and results shift when fossils are incorporated.
A third, related but often unstated assumption for retrospective analyses from extant species is that a focal clade is today at its maximum morphological breadth; this is necessarily the case when phylogenies contain only extant species, but is patently false for many clades having a reasonable fossil record, from oysters to cephalopods to elephants to horses to hominins. The extinct forms are often not simply extensions along existing morphogenetic lines but variations that might seem highly improbable given today’s representatives, for example giant ground sloths (terrestrial and aquatic), rainforest-dwelling carnivorous kangaroos, sharks with coiled tooth arrays, uncoiled or spiny nautiloids, echinoids with periscope-like extensions, and so on (see Jablonski, 2020 for references; also Stubbs et al., 2013 and Melstrom & Irmis, 2019 on insectivorous and herbivorous Crocodylomorpha). Even the quintessentially static lineage, the horseshoe crabs, has undergone bursts of phenotypic diversification that pushed beyond their current limited repertoire, corresponding to invasion of new habitats (Bicknell et al., 2022; Lamsdell, 2016). Wagner (2010) elegantly makes this general point by comparing Cambrian and present-day arthropods, which exhibit similar disparities (despite the far more limited Cambrian sample!) with relatively little overlap in morphospace; Carboniferous arthropods (ca 320 Myr old) bridge between them temporally and morphologically, but also add further disparity.
Observations in the Fossil Record
Paleontological analyses pertaining to evolvability labor under a different set of strong assumptions. Sampling and preservation can distort or even generate apparent patterns, although increased understanding of such potential biases and resulting methodological advances can reduce their impact. Only post-embryonic, phenotypic data are available for most extinct taxa, so that the developmental and genetic underpinnings of observed contrasts, and the intrinsic vs extrinsic controls on differential behavior of clades, must be inferred. Particularly challenging is the assessment of negative evidence (also an issue for neontological data of course), and of the role of intrinsic and extrinsic factors in determining vacancies or boundaries of a clade’s morphospace. Some vacancies are long-standing and phylogenetically localized, and thus may represent a lack of developmental capacity, at least for the clades presented with these opportunities (Jablonski, 2020; Vermeij, 2015). Others may reflect extinction and insufficient time to re-occupy vacated morphospace (consider mammalian body sizes in the Americas, although humans have surely now blocked that evolutionary route). Morphospace occupation also can be limited by pre-emptive occupation or later, displacive conquest of portions of the space by competing clades. Displacive competition seems to be scarce at macroevolutionary scales, but pre-emptive, incumbency patterns or priority effects are evidently more common (see Jablonski, 2008b, 2017b; Benton, 2009; Tilman & Tilman, 2020; and Tomiya & Miller, 2021 for a study that may find both effects). Other negative interactions, such as predation and parasitism, can promote or impede phenotypic or taxonomic diversification; as can positive interactions such as mutualism, and either type can sometimes increase extinction probabilities (see Vermeij, 1987; Jablonski, 2008b; Hembry & Weber, 2020). Again, comparative analyses of clades presented with similar opportunities can control for some of these uncertainties, and temporal and spatial paleo-data can be especially valuable, with insight into a fuller range of phenotypes accessible to a clade than may be seen today, and into potential interactions: clades cannot impede one another if they did not co-occur.
Despite these drawbacks and complications, many analyses do suggest among-clade and temporal differences in evolvability, with macroevolutionary consequences. Some of these are discussed below.
Features Enhancing Evolvability of Clades
Modularity
The developmental property most often held to be associated with evolvability is modularity. The general view has been that greater modularity enhances evolvability (e.g. Love et al., 2021; Vermeij, 1974, 2015, as “versatility”, which he associates with modularity in the later paper; Wagner & Altenberg, 1996). However, many different types of modules are recognized, i.e., functional, developmental, genetic, and evolutionary modules (references in Jablonski, 2017a), and we lack clarity on how they are related, with mixed results on the positive, negative, or negligible relation between the strength of modularity and macroevolution (Rhoda et al., 2021 and references therein). For modularity to enhance evolvability, the intrinsic structure of modules—i.e. genetic or developmental modules—must be configured along viable lines, which need not be the case (e.g. Pavlicev & Hansen, 2011), and must align with extrinsic selection; otherwise trait covariation within modules can instead impede evolution. This contingent aspect of modularity would seem to disallow generalizations, and macroevolutionary predictions become difficult, although retrospective understanding of a role for modularity in specific cases is not a trivial insight. However, the ubiquity of mosaic evolution, and more broadly, of inconsistent character transformations across phylogenies (Jablonski, 2017a), indirectly supports the view that evolution is often facilitated by the ability of traits to change independently.
Further, among-clade differences probably exist: arthropods seem to be masters of modularity, not just in terms of dissociating morphological modules for independent growth and transformation (e.g. Nijhout & McKenna, 2017), but perhaps also at the molecular level. For example, arthropods apparently more readily deploy the Distal-less pathway in new locations to generate novel structures (horns, wings, pigmentation eyespots; see Shubin et al., 2009; Murugesan et al., 2022) than do tetrapods, whose lack of wings sprouting from their backs instead of supported by forelimbs has often been used to exemplify developmental constraint (Erwin, 2007; Jablonski, 2017a; Losos, 2011). The greater ability of arthropod Distal-less to generate novelty relative to its vertebrate homolog Dlx may derive from stronger modularity at two levels: organismal, with more overt segmentation in arthropods, and genomic, with the arthropod pathway largely dedicated to regulating outward growth but a much more extensive pleiotropic repertoire for vertebrate Dlx, which is involved not just in the early development of limbs, but in the placenta, forebrain, branchial arches, and other tissues (Panganiban & Rubenstein, 2002; Sumiyama & Tanave, 2020).
A related view sees evolvability as a positive function of the dimensionality of form (Vermeij’s, 1974 argument), which need not be directly related to modularity per se: limpet shells can be described by fewer mathematical parameters than can helically coiled shells with complex apertures, and thus have lower dimensionality, but different snail lineages have not been analyzed from an evolvability perspective (for more on the positive associations between dimensionality and the rate or extent of diffusion in morphospace, see Foote, 1991:129; Pie & Weitz, 2005:E9). In a sense this is a “degrees of freedom” hypothesis—more components mean more avenues to evolve along, or, in Vermeij’s (2015) view, for alleviating functional tradeoffs. For example, Le Maître et al. (2020) argue that incorporation of jaw bones into the mammalian middle ear increased the number of genetic and developmental factors involved in the auditory system, and so enhanced its evolvability relative to the simpler reptilian and avian ears. The positive effects of complexity on evolvability can also be viewed in functional terms: when more traits contribute to performance, evolution is more often able to circumvent potential tradeoffs, as in the ability of centrarchid fishes to combine high suction forces with large gapes (Holzman et al., 2011; see also the general discussion and novel tradeoff model of Polly, 2020). Relating the shifting combinations of functional traits to developmental modularity remains challenging but may help to explain among-clade and among-trait differences in lability.
Perhaps evolvability is actually greatest at intermediate strengths or extents of phenotypic modularity, and perhaps at intermediate phenotypic dimensionality (Hansen, 2003: p. 87; Pie & Weitz, 2005). Too much fine-grained modularity may decrease the potential for substantial evolution by requiring many small mutations that separately affect each of many independent traits (a “cost of complexity,” Orr, 2000), whereas too little modularity (roughly but imprecisely equivalent to too much integration) may approach a state of universal pleiotropy, where virtually any genetic change adversely affects other aspects of the phenotype. Intermediate levels of morphological integration are associated with the greatest degrees of evolutionary divergence in artiodactyls (Haber, 2016), but more work is needed to assess this hypothesis.
As in many other aspects of macroevolution, contingency is a factor when considering the potential role of trait covariation in evolvability. If rate and net distance traversed in morphospace is the evolvability measure, as suggested here, the covariation structure imposed by morphological integration—as noted, not strictly the antithesis of modularity but useful in this context—can enable more rapid and extensive evolutionary change in certain directions than would emerge from strictly isotropic or unbiased variation (Felice et al., 2018; Goswami et al., 2014; Jablonski, 2020; Love et al., 2021; Uller et al., 2018). Thus, in the special circumstance when selection (i.e. an opportunity) is aligned with such (viable) lines of genetic least resistance in Schluter’s sense, integration rather than modularity can promote greater evolvability. As Goswami et al. (2014) put it, “a more modular system will explore a greater volume of a morphospace than a more integrated one … but it will not evolve phenotypes as maximally disparate as a highly integrated system that forces all variation along a relatively narrow trajectory;” see also Evans et al. (2021) on instances where highly integrated traits appear to have been most evolvable. This view also implies that the evolvability of a clade will change over time as an indirect byproduct of shifts in morphologic integration, and more generally in covariance structure, shifts in strength or configuration (see Wagner, 2022).
These contingencies are also reflected by the finding that developmental simplicity evidently promotes evolvability in some situations, and we need a deeper analysis of how the predominant views on phenotypic modularity or dimensionality relate to the fact that bird beaks have more broadly diversified when underlain by a simple coordinate system determined by a few key regulatory pathways, as in certain finch clades (Abzhanov et al., 2006; Mallarino et al., 2011; Tobias et al., 2020). Modularity as manifest in mosaic evolution has been a major theme in beak evolution since the Cretaceous (O’Connor et al., 2020), providing many opportunities for comparative analyses of clades through their histories.
Of course, the diversification of the bird beak is also enabled in part by its status as a module relative to other parts of the body, but Navalón et al. (2020) argue that greatest disparity, albeit in restricted directions in morphospace, has been achieved in those clades having a high degree integration of the beak with the rest of the skull, just predicted by Goswami et al. (2014); see also Hedrick et al. (2020) on bat cranial evolution. Central to these ideas from a macroevolutionary perspective is the still-open question of the long-term stability of genetic and phenotypic modularity (see Urdy et al., 2013; and for a mix of stability and lability in archosaur crania see Felice et al., 2019), and how to operationally distinguish modules maintained by intrinsic factors resistant to change, from those maintained by selection and thus readily altered at these large scales. Experimental analyses can be useful, but may give different results from natural populations (Klingenberg, 2008, 2014). Here too, retrospective macroevolutionary analyses of clades with demonstrable present-day differences in modularity would be a powerful merger of paleontological and neontological data. Ideally, we could compare two clades differing in modularity but presented with a similar opportunity, such as survival of a mass extinction, or arrival in a relatively unoccupied archipelago or larger landmass. However, simply enriching our picture of the permanence or lability of apparent modules using the fossil record can be revealing. For example, tetrapod forelimbs and hind limbs are often portrayed as a module, later broken by bipedalism (Young et al., 2010), so that coordinated enlargement or reduction of fore- and hind-limbs might be the expected in most instances. However, even in the Carboniferous Period, not long after tetrapod origins, limb reduction and loss occurs forelimb-first (Mann et al., 2022), a dissociation also seen at the origin of snakes (e.g. Zaher et al., 2009), and hindlimb-first reduction occurs in many other groups (see Mann et al., 2022, for references).
Given the array of skeletal types that constitute almost all of the fossil record, we might ask whether developmental and evolutionary modularity—and thus potentially evolvability—differs between body plans involving many-element, articulating skeletons such as vertebrates, echinoderms, and arthropods, and those having just one or two discrete elements and accretionary growth, such as corals, mollusks, and brachiopods (see for example Edie et al., 2022). The remarkable range of molluscan shell shapes (that is, scaphopods, nautiloid cephalopods, chitons, snails, and bivalves) and ornamentation patterns suggest exquisite local control in the sheet of tissue that generates those shells, and Herlitze et al. (2018) suggest that the evolvability of the molluscan shell may ultimately derive from a high degree of “spatial modularity” in distinct sets of genes and cell lineages within that tissue. The unaddressed question is whether the extra level of morphogenetic control and interaction afforded by articulating skeletons creates a correspondingly enlarged evolvability at macroevolutionary scales.
An even more profound difference between clades that could be viewed from the modularity/evolvability standpoint involves lineages that sequester the germ line early, versus the plants and clonal colonial animals that sequester the germ line late and so can incorporate somatic mutations into gametes (Schoen & Schultz, 2019 and references therein; Simpson et al., 2020; Yu et al., 2020). With late sequestration, each plant bud or animal zooid is potentially both developmental and evolutionary module, so that novel variants can originate within the colony and propagate both sexually and asexually, conceivably increasing clade evolvability relative to early-sequestration clades, with the added benefit of far greater ability to adjust reproductive allocation to immediate conditions (Hiebert et al., 2021). Still unclear is whether the short-term benefits of promoting beneficial mutations via intraorganismal selection, and more generally by providing greater variation to local populations (e.g. Folse & Roughgarden, 2012), translates into significant differences in evolvability at macroevolutionary scales, but the fact that early sequestration is the derived state for both plants and animals (e.g. Buss, 1987), suggests that they generally do not, in many animal groups, as does apparent suppression of somatic mutation rates in large, long-lived plants (Orr et al., 2020). At the very least, clonal animal colonies go beyond the modularity seen in plants to add a potential level of selection to the biological hierarchy (Buss, 1987; Simpson, 2012; Simpson et al., 2020). Little-explored tradeoffs may exist between colony- and zooid-level diversification, as seen in the evolutionary differences between corals (with essentially monomorphic zooids but an extraordinary range of colony forms including solitary individuals) and bryozoans (richly polymorphic at the zooid level but lacking comparable colony sizes or diverse solitary forms) (Lidgard et al., 2012; Schack et al., 2019; at least 10 phyla have evolved polymorphic colonies, but most colonial species lack polymorphic zooids, Hiebert et al., 2021).
The notion that clonal colonial organisms or plants emphasizing vegetative reproduction might be more evolvable might seem to contradict a widely (though not universally) accepted case of higher-level selection for evolvability: the pervasiveness of sexual reproduction across the tree of eukaryotic life. The Red Queen hypothesis for the maintenance of sex, e.g. parasite-mediated selection for the continual production of novel phenotypes, defines a process playing out at the population, species and/or clade level (Van Valen, 1975; Stanley, 1979: pp. 213–227; Nunney, 1989; Sterelny & Griffiths, 1999: pp. 208–210; Hansen, 2011; de Vienne et al., 2013), and thus is very much a macroevolutionary hypothesis. But there need be no contradiction here: species in most eukaryotic clades that reproduce asexually or parthenogenetically are also capable of sexual reproduction. Testing a macroevolutionary hypothesis of the consequences of evolvability as imposed by sex could involve asking whether lineages in which sexual reproduction is rare or involves a limited number of individuals are less prolific phenotypically than lineages in which sex is the norm. Because sexually produced individuals or colonies can be distinguished from asexually produced ones in several well-fossilized groups (foraminiferans, corals, bryozoans), this question could be addressed empirically. More challenging to address on macroevolutionary scales is the hypothesis that among-clone (and therefore among-clade) selection for evolvability might operate through the structure and interaction of gene regulatory networks, as proposed by Woods et al. (2011) in their long-term E. coli experiments. If some genetic attributes consistently impose a greater potential for further adaptation, as Woods et al. put it, then retrospective tests may be feasible. Such tests may circle back to modularity as noted above, but that explanation does seem to apply to E. coli (for other potential prokaryote examples, see Payne & Wagner, 2019).
Ontogenetic Allometry or Multiphase Life Cycles
As noted above, development can impose covariation patterns enabling more rapid and extensive evolutionary change in certain directions than would emerge from strictly isotropic or unbiased variation, so that phenotypic integration can enhance evolvability if aligned with selection. This largely theoretical view has been supported by a few analyses, but may find its richest macroevolutionary potential in clades that undergo strong changes in form during ontogeny, as continuous variation in ontogenetic allometry—which can be viewed as a form a morphological integration (Hallgrímsson et al., 2019)—or discontinuously in multiphase life cycles. As long recognized (e.g. Gould, 1977), such clades have often evolved along ontogenetic trajectories via heterochrony, i.e. evolutionary changes in developmental timing, and in at least some cases traverse significantly greater volumes of morphospace than clades with lesser allometries or more direct development. Examples include the origin of sand dollars (Smith, 2001) and brittle stars (Thuy et al., 2022); and evolution within canids (e.g. Geiger et al., 2017; Machado et al., 2018: p. 1413; domesticated dogs are not simply paedomorphic wolves but the extreme disparity of dog breeds involves heterochrony; and for a broader overview see Sánchez-Villagra et al., 2017), snakes (Esquerré et al., 2017), non-avian dinosaurs (Chapelle et al., 2020), kingbirds (Fasanelli et al., 2022), and angiosperms (Armbruster, 2022), all exploiting ontrogenetic allometries; and most famously regarding indirect development, extant and fossil salamanders that retain larval traits, with modularity clearly a critical part of this capability (see Johnson & Voss, 2013; Urdy et al., 2013; Fabre et al., 2020).
The frequency of heterochronies that carry larval traits into adult phenotypes is unclear, salamanders aside, and of course some paedomorphic salamanders have instead drawn on later ontogenetic changes (Alberch, 1980; Wake, 2009). Indirect-developing marine invertebrates contrast with salamanders in showing few cases of heterochrony across the metamorphic event, but see Teichert and Nützel (2015) and Tajika et al. (2018), on the multiple paedomorphic origins of pelagic gastropods, arguing first for an oceanographic trigger and then for a later opportunity created by the end-Cretaceous mass extinction (but why didn’t co-occurring bivalves or sea urchins rise to either occasion, so to speak)? Flowing in the opposite direction, and likely much deeper phylogenetically, Marlow (2018) argues that most marine feeding larvae represent heterochronic shifts from adults to larvae, as adaptations to prolong pelagic durations via enhanced feeding & digestion.
Insects that undergo complete metamorphosis might also be viewed in this light, although most analyses emphasize the ecological separation of larva and adult (with the pupa a non-feeding transitional stage) and resulting taxonomic diversification rather than phenotypic evolvabilty (e.g. Rainford et al., 2014; Yang, 2001). As in mollusks and echinoderms, metamorphosis may be such an extreme event in insect development that larval characters are unlikely to carry over into adults (e.g. Trueman, 2019; but see McMahon & Hayward, 2016, who find a few, rare, examples). As with salamanders vs frogs, hemimetabolous insects may be more likely to draw on, but perhaps also be more constrained by, their ontogenetic trajectories than holometabolous ones, but the net evolutionary consequences have not been assessed (see Moore & Martin, 2021; Galis, 2022). Also unknown is whether clades differ in their ability to change scaling relationships and thus ontogenetic allometries (e.g. Tsuboi et al., 2018; Voje et al., 2014): localized scaling shifts could propel developmental modules into new regions of morphospace (Nijhout & McKenna, 2017), but among-clade frequencies have not been evaluated. Some data on sticklebacks suggest that allometric patterns fade as evolutionary constraints over 1–2 Myr (Voje et al., 2022), just as Hunt (2007) found for the influence of (co)variance patterns on speciation directions; Esquerré et al. (2017) also found ontogenetic allometries to be “highly evolvable”, but the timescale is less clear; see also Natale and Slater (2022) on shorebirds.
Some multispecies trends are reported in the fossil record as successive directional shifts toward increasingly paedomorphic or peramorphic states (paedomorphoclines and peramorphoclines, respectively, reviewed by Jablonski, 2020; Lamsdell, 2021; McNamara, 2012). Such patterns are intriguing as potential cases of heightened evolvability in particular directions, but most require formal phylogenetic analysis and more detailed morphometrics to confirm the stepwise, anagenetic dynamics portrayed in such studies. Even if clade topologies prove to be more complex than first inferred, the observation that clades show greater net evolvability in directions predictable from ancestral ontogenies is likely to be robust, whether in cladogenetic or anagenetic mode (Jablonski, 2017b); more detailed macroevolutionary comparisons to relatives showing less ontogenetic variation would also be valuable.
Novel Traits
Evolutionary novelty in the broad sense often seems to increase evolvability, by creating new features for further variation, and allowing clades to access new adaptive zones (Simpson, 1944): the origin of limbs, lungs, the amniote egg, and feathers are certainly associated with an expansion in the morphological disparity (and taxonomic diversity, and functional repertoire) of the clades bearing them. However, we have surprisingly few robust examples of this key-innovation phenomenon, in which a novel feature directly triggers diversification (see Rabosky, 2017; Martin & Richards, 2019; Erwin, 2021a for recent catalogs and critiques of the many definitions of “key innovation”). Many putative key innovations have proven to be part of a chain of derived characters, or associated with “key opportunities”—i.e. extrinsic events—prior to phenotypic expansions (see Donoghue & Sanderson, 2015; Stroud & Losos, 2016; Jablonski, 2017a). Such contingencies are most clearly seen in macroevolutionary lags, the geologically long interval between the inception of a novelty or clade and its taxonomic or phenotypic diversification (Jablonski & Bottjer, 1990), which appear to be widespread or even the general rule (Jablonski, 2017a; Halliday et al., 2019; Kröger & Penny, 2020; Ramírez-Barahona et al., 2020; Simões et al., 2020, Erwin, 2021a; see Near et al., 2012 for one of many possible examples from time-calibrated molecular phylogenies; and Moharrek et al., 2022, whose work confirms Jablonski & Bottjer, 1990 and Patzkowsky, 1995 on bryozoans without citing them). Such lags can be valuable analytical tools, providing a novel framework for evaluating intrinsic and extrinsic factors, by pinpointing an apparent increase in evolvability at a specific point in a clade’s history. Many will likely prove to involve diversifications in the aftermath of mass extinctions or other environmental triggers (see Slater, 2013 on the change in evolutionary mode in post-Cretaceous mammals; Erwin, 2021b on the Cambrian explosion of metazoan phenotypes relative to the divergence of the major clades), but some will involve intrinsic phenotypic traits that have compounded to promote diversification intrinsically (e.g. both Jablonski et al., 1997 and Taylor & Waeschenbach, 2015 on bryozoans). The cited examples notwithstanding, macroevolutionary lags are generally tracked using taxonomic diversity, and more analyses are needed that treat them in morphospace and functional variety (as in Folk et al., 2019).
We do not know how often evolutionary novelties in the strict sense—i.e. a trait lacking a homolog in the ancestor (Wagner, 2014)—also fail to serve as diversification triggers. As these true novelties often define clades, analyses of lags will need to operate across broad evolutionary trees, but effects seemingly imposed by intrinsic constraints and their removal or absence may also present a useful set of test cases. Here too, rigorous macroevolutionary analyses are scarce. Mammals are highly constrained in the number of cervical vertebrae (Galis, 2022; Galis et al., 2018), but this constraint has been circumvented elsewhere in tetrapod phylogeny as seen in sauropod dinosaurs, plesiosaurs, and other Mesozoic clades, and in long-necked birds today (Müller et al., 2010; Taylor & Wedel, 2013). The question, however, is not whether, for example, Anatidae (e.g. swans) have diversified taxonomically more than, say, Felidae (cats), but whether the constraint on cervical vertebrae has impaired mammalian functional or morphological evolution relative to clades bearing no such constraint (e.g. Marek et al., 2021). Thus, the first research target should probably be identifying where and how often in tetrapod phylogeny that constraint was broken, or acquired (e.g. Müller et al., 2010).More generally, a systematic inquiry is needed into the underlying basis of within-clade increases in evolvability as defined here: how often do they actually entail changes in the genotype–phenotype map, or restricting of genetic or developmental modularity? (The literature is large but inconclusive, for discussion and references see Hansen, 2006; Wagner, 2014; Jablonski, 2017a; Erwin, 2021a.)
Another intriguing modification of development, little-studied from the standpoint of evolvability, is the breaking of bilateral symmetry. Examples occur throughout plant and animal phylogeny, by a variety of developmental mechanisms (Palmer, 2004). Some changes are subtle, or associated with developmental “noise.” Such “noise,” such as fluctuating asymmetry, has been hypothesized as an indicator of weak canalization and therefore of enhanced evolvability (see Webster, 2019), although more work is needed on this idea. No one has evaluated whether the ability to adopt fixed asymmetry enhances evolvability. Bivalve mollusks are a system that would reward such an analysis (Fig. 2). Most species are bilaterally symmetrical, aside from small developmental adjustments allowing interlocking, hinged valves (Moulton et al., 2020; recall that the plane of symmetry lies between the two valves, not down the midline of a single valve), and this is the ancestral state (e.g. Audino et al., 2021; Waller, 1998). However, some bivalve clades have strongly diverged from bilaterality, including the extinct, perhaps photosymbiotic, rudists, which evolved a conical-cylindrical right valve and a cap-shaped left valve, among other configurations (Skelton, 1985). Oysters, spiny oysters, scallops, and others have also shed bilateral symmetry in impressive ways (Harper & Checa, 2020; Nicol, 1958; Sherratt et al., 2016), with extinct oysters showing a much wider range of shell geometries than extant species, including planispiral, helical and conical forms. As many of these lineages are in the Order Pteriomorphia, the question arises whether this clade weakened bilateral patterning early in bivalve history and then could adopt asymmetry according to later opportunities or pressures, and thus had greater evolvability than related bivalve clades. An alternative is that bivalves were never strongly constrained to bilateral symmetry so that the current distribution of this trait simply reflects the ecological history of different clades; these alternatives may be testable phylogenetically in the fossil record, and perhaps via experimental manipulation of extant species.
Breaking bilateral symmetry in bivalves. A Cretaceous rudist Radiolites angeiodes showing disparate left (now upper) and right (now lower) valve (after Skelton, 1979, used by permission). B Eocene Caestocorbula praeviator, living within sediment with larger and more heavily ornamented left valve downward (from Beu & Raine, 2009). C extant scallop Cyclopecten hoskynsi, left valve above, right valve below (from Dijkstra et al., 2009). D Coiled Jurassic oyster Gryphaea arcuata, living with left valve downward on soft seafloor (from Seilacher, 1984). E Helically coiled Cretaceous oyster Ilymatogyra arietina, living with left valve downward on soft seafloor (from Roemer, 1862)
A shift from radial to bilateral symmetry is associated with a striking contrast in apparent evolvability in sea-urchin history (Fig. 1). The ancestral condition is radial, and the survivors of the end-Paleozoic mass extinction inherited that state, continuing to evolve as the group informally termed regular echinoids; they gave rise to many species but remained confined in morphospace. However, one lineage diverged to become the irregular echinoids, a bilaterally symmetrical, burrowing clade that eventually split into two branches typified respectively by heart urchins and sand dollars. The regular and irregular echinoids each contain ~ 500 extant species, but the irregulars have explored a much broader range of morphospace (Hopkins & Smith, 2015; see also Boivin et al., 2018, on the expansion in functional diversity in irregulars). Understanding the developmental basis of this contrast, including a potential change in modularity (López-Sauceda et al., 2014; Saucède et al., 2015), and then testing an intrinsic evolvability hypothesis against alternatives—bilateral symmetry as a key innovation, or ecological opportunities afforded by adoption of the burrowing, deposit-feeding habit (and ultimately a suspension-feeding one in sand dollars), would create an exceptional model system for exploring macroevolutionary issues. One factor may be a profound change in growth processes near the origin of irregulars (Smith, 2005): their plates predominantly grow in place throughout ontogeny (as opposed to growth by a combination of plate growth and insertion in regulars), making it easier to differentiate the upper and lower surfaces of the test, and thus to become burrowers, or, as in sand dollars, to use the upper surface as a feeding sieve. Shifts from radial to bilateral symmetry may also promote diversification in angiosperms, separately or in combination with other traits (O’Meara et al., 2016; but see Reyes et al., 2016, and Vamosi et al., 2018). However, this effect has only been evaluated in terms of species richness and not phenotypic evolvability, and comparative analysis of floral evolution in morphospace according to floral symmetry would be a valuable next step.
The converse of a macroevolutionary lag is the dead-clade-walking (DCW) phenomenon, where a clade suffers a sharp decline, e.g. at a mass extinction, and then persists for some time without re-diversifying (Jablonski, 2002). Like macroevolutionary lags, such clades appear to be widespread (Barnes et al., 2021), and just as lags appear to signal a belated gain in apparent evolvability, the DCW pattern may signal a clade’s loss of evolvability, or more precisely, are potential natural experiments in the loss of traits thought to promote evolvability, for comparison to clades that retain those traits. As with lags, many of the DCW’s may actually involve extrinsic factors, such as limits imposed by competitors or predators in the post-extinction world, but analyses are lacking, and we cannot rule out the loss of an intrinsic evolvability-promoting feature, at the organismal level or higher (e.g. genetic population structure). DCWs have only been analyzed taxonomically, so that we still need to know if they are phenotypically or functionally static after their bottleneck, and thus provide a vehicle for directly testing hypotheses on drivers of evolvability. On the other hand, if they shift significantly through morphospace despite low taxon numbers, they could not be viewed as suffering diminished evolvability in the sense used here.
Another way to reduce evolvability is by entry into certain modes of life that evidently involve irreversible commitments, as in hypercarnivorous mammals (Martin, 2019; Van Valkenburgh, 2007), and as suggested for uniparental reproduction in animals (e.g. Fujita et al., 2020). We do not know whether such contrasts represent the idiosyncrasies of specific functional groups and their surrounding adaptive landscapes, or more general intrinsic shifts associated with “evolutionary ratchets”, for example involving the number and integration of traits involved in the adoption of a given mode of life.
Genome Size
For plants, genome size, and specifically whole-genome duplication (WGD) related to interspecific hybridization and allopolyploidy, has been tied to evolvability. Allopolyploids can create unique amalgams of parental phenotypes and generate novel features (e.g. Alix et al., 2017; Soltis et al., 2014), so that plant clades more prone to allopolyploidy, and/or with more WGDs in their history, should traverse or occupy more morphospace than other clades. This prediction is evidently met at a broad scale among the major angiosperm clades, with core eudicots, arising with a genome triplication, occupying a greater volume of morphospace than basal eudicots, monocots, or magnoliids (Clark & Donoghue, 2018; conifers show less robust but consistent patterns). Much more work is needed to test the potential mechanistic link (see for example Zenil-Ferguson et al., 2019, and the controversy regarding polyploidy and taxonomic diversification, references in Bowers & Paterson, 2021), and to understand how these results fit into the lack of an association between WGD and disparity in mosses and horsetails (Clark & Donoghue, 2018; Clark et al., 2019). These results should also be reconciled with the fairly constant overall disparity of angiosperms and several of their subclades since the Cretaceous (Oyston et al., 2016; measured differently from the preceding study), despite a phylogenetically widespread set of WGDs that evidently occurred near the end-Cretaceous extinction (see Levin, 2020). The ability to estimate ploidy levels from fossil material provides an excellent vehicle for more rigorous tests of this hypothesis without reliance on ancestral character-state estimation from extant species (e.g. Lomax et al., 2014; Masterson, 1994; McElwain & Steinthorsdottir, 2017).
The macroevolutionary role of genome size in animals is unclear. Ancient WGDs have been associated with early taxonomic and morphological diversifications in vertebrate and invertebrate clades (e.g. Conant, 2020; Liu et al., 2021). However, we need more direct evidence on how those apparently exceptional deep-time events affected phenotypic evolution—the potential role of post-duplication neofunctionalization and subfunctionalization is much-discussed—and whether later changes in genome size within smaller clades have had similar apparently positive effects, particularly in fishes and amphibians, where they are best-documented (Mable et al., 2011; Aase-Remedio & Ferrier, 2021). The multiple rounds of WGDs in fishes still need investigation from the evolvability standpoint, with two WGDs near the origin of vertebrates, another WGD near the origin of teleosts but well-separated from the modern diversification of the group in an apparent macroevolutionary lag (Davesne et al., 2021; Glasauer & Neuhauss, 2014), and younger events, e.g. near the origin of salmonid teleosts and now in the process of rediploidization (Lien et al., 2016), also with an apparent lag between WGD and diversification, in this case ~ 40–50 Myr.
Although increases in genome size may promote paedomorphosis by slowing development (with implications discussed above), they may tend to damp evolvability in animals, in apparent contrast to plants (Kraaijeveld, 2010; Lertzman-Lepofsky et al., 2019; Womack et al., 2019). What animals do share with plants is the potential to track genome size directly in the fossil record through measurements of cell dimensions in, for example, well-preserved bone or shells, a largely neglected opportunity (but see Thomson & Muraszko, 1978; Conway Morris & Harper, 1988; Ushatinskaya & Parkhaev, 2005; Organ et al., 2007, 2011; Hunt & Yasuhara, 2010; Davesne et al., 2021). Of course, genomes can enlarge for reasons other than duplication, and one of the most interesting potential directions for macroevolutionary investigation in this area is the relative impact of transposon proliferation and WGD on clade survivorship and diversification, which might be assessed retrospectively when phylogenetic analysis shows a constant ploidy level but fossil data indicate shifts in genome size. There are many ideas on the evolutionary role of mobile elements, some of them plausible, including the potential for cross-level conflicts, but the macroevolutionary impact of among-clade differences in transposon content—active or not—remains uncertain.
Evolvable Traits
As noted above, individual phenotypic traits might differ in evolvability, and to the extent that they facilitate the expansion of clades in morphological or functional range, might impart greater overall evolvability to the clade. Many attempts have been made to frame macroevolutionary generalizations for trait evolvability. Developmental burden or entrenchment, where other traits are contingent on the development of the focal trait, is thought to limit evolvability of the focal trait (e.g. McGhee, 2011; Riedl, 1978; Wimsatt & Schank, 2004), and some authors have attempted to formalize such burden in terms of position within gene regulatory networks: traits determined at the tips of such networks should be freer to evolve than traits controlled at higher levels (Erwin & Davidson, 2009). This idea is appealing, but it may be difficult to sustain. In a multilevel system, gene-network conservatism need not be reflected in phenotypic evolvability—consider the extraordinary conservation of networks patterning the heart, eye, or appendages, that have been redeployed in the service of evolutionary novelty (e.g. Wagner, 2014), and conversely, developmental system drift, in which morphology is conserved while the underlying developmental pathways change (DiFrisco & Jaeger, 2021; True & Haag, 2001; Wagner, 2014). Raff (1996: pp. 316–318) goes further, arguing that the hourglass model of development, with conservatism in form strongest at mid-embryogenesis, essentially invalidates most concepts of burden. This model does suggest that the most upstream regulatory genes are not maximally conserved, but more work is needed to test the possibility that the apparent phenotypic bottleneck in development shifts this form of constraint and its impact on evolvability downstream genetically and developmentally, but does not refute it altogether—gene expression also diverges least in the phenotypically defined hourglass bottleneck (Wagner, 2014; see also Piasecka et al., 2013; Valentine & Marshall, 2015). Translating such observations to a concept of burden may still be undermined by direct evidence, still not plentiful, that differences among related species can be underlain by genetic changes deep in the regulatory hierarchy (Lavoie et al., 2010; Streelman et al., 2007). Many aspects of body plans do seem highly resistant to change—hence the anisotropic variation that typifies developmental bias—but the geometry of regulatory pathways and their recursive circuits may be too complex for simple generalizations on evolvability (Deline et al., 2020; Erwin, 2021a: p. 7). As with phenotypic modules, modular organization of developmental processes may help to circumvent burden on specific traits, with the more evolvable ones being those whose regulatory factors are structured such that small developmental changes produce significant shifts in form, as Jernvall (2000) suggested for size and number of cusps in mammalian teeth (see also Burroughs, 2019; Couzens et al., 2021).
Another attribute that might enhance trait evolvability is plasticity, the ability to produce different phenotypes in response to environmental cues. In principle, the broader the reaction norm, the greater the opportunity for genetic assimilation or accommodation of alternative traits or trait values (e.g. West-Eberhart, 2003; Pfennig., 2021; Levis & Pfennig, 2021). Three considerations are necessary. First, direct evidence for different levels of plasticity among diversifying clades will be needed to test for associated differences in evolvability, but separating plasticity from genetic variation in fossil populations is not straightforward (Webster, 2019; see Lister, 2021 for a more optimistic view). Second, plasticity is not always adaptive, and becomes increasingly maladaptive with increasing environmental stochasticity (references in Pfennig, 2021). This complicates predictions for evolvability, but opens up other interesting issues with respect to the potential effects of destabilized past and current environments. Third, plasticity depends on environmental cues, so that if plasticity does play a significant macroevolutionary role, then the altered environmental contexts frequent at large temporal and spatial scales, such as geographic range expansions, could change the evolvability of a lineage (Love, 2003). From a macroevolutionary standpoint, plasticity is another unquantified potential source of variability, and likely another mechanism for presenting biased variation to selection and other evolutionary processes (Parsons et al., 2020).
The functional roles of traits may also affect their evolvability, and ultimately impose evolvability differences among clades. Tradeoffs are often invoked for limitations on disparity, but can be surprisingly elusive for the morphological traits most accessible to macroevolutionary analyses. As noted above, increasing the number of traits collectively engaging in a given function may mitigate the effects of potential tradeoffs. However, single traits that serve multiple functions may be less evolvable, i.e. generate less disparity, than traits serving fewer functions. For example, performance tradeoffs may limit disparity in aquatic turtle shells, which must both resist loads and reduce drag, relative to the shells of terrestrial turtles (Stayton et al., 2018; and Stayton, 2019 for an additional performance demand). Perhaps trait-level tradeoffs promote the parcellation of traits, i.e. their evolutionary partitioning into several functional modules, via the break-up of developmental modules (see Wagner & Altenberg, 1996).
Elevated Speciation Rates
Over geologic timescales, most species tend to be morphologically static (i.e. oscillate within limits) or nondirectional over their histories, affording speciation a potential role in the extent and direction of morphospace occupation for many clades (e.g. Gould, 1982; Hunt, 2007; Jablonski, 2017b; and from a very different perspective, Gorné & Diaz, 2019). Some authors include high speciation rates in their definition of evolvability (e.g. Hedrick et al., 2020) although I argued against such a broad definition above. In any case, we can ask whether clades having higher speciation rates for intrinsic reasons—owing to traits that increase the probability of reproductive isolation (see Jablonski, 2008a)—have higher rates or extents of net morphospace occupation. (Such analyses will not be circular if performed with care, even in the fossil record where speciation is necessarily recognized phenotypically, because the critical variable is net differences in morphospace occupation). A rough correlation between speciation rate and morphological change is seen for many clades at various points in their histories, albeit with considerable heterogeneity and an array of counter-examples (Stanley, 1979; Rabosky et al., 2013; Crouch & Ricklefs, 2019; Cooney & Thomas, 2021; and many more citing and cited in these publications; see below for temporal changes such as early bursts).
The potential association between speciation and morphologic change is relevant to evolvability for at least three reasons.
-
(a)
Speciation may tend to occur preferentially in the direction of intraspecific (co)variation (Hunt, 2007; Love et al., 2021; Polly & Mock, 2018), providing a potential link between standing variation and both developmental bias and macroevolutionary evolvability, with high-speciation clades moving more rapidly across morphospace per unit time, and more efficiently in that fewer species go in the opposing direction over the course of the trend—Gould’s (1982) “direction bias” in clade dynamics (see also Jablonski, 2020). (Similar results are reported, mostly over shorter time scales—perhaps just a few generations—for patterns of genetic (co)variance and phenotypic divergences, but as noted above macroevolutionary predictions may be undermined by the complex, nonlinear relationships between genotype and phenotype often described as G-matrices and P-matrices (Hallgrímsson et al., 2022; Milocco & Salazar-Ciudad, 2022; but see Saltzberg et al., 2022 for a possible counter-example). The initial directionality imposed by standing phenotypic variation is longer-lived but tends to fade after a few million years, or is disrupted by extrinsic events such as climate shifts (Brombacher et al., 2017; Guenser et al., 2019; Hunt, 2007; Schluter, 1996). Such instability seems to limit the macroevolutionary role of this phenomenon, but the several potential mechanisms behind it have not been explored. Further, the orientation and eccentricity of larger‐scale multivariate variation is stable at higher levels within larger clades, and is also associated with evolutionary trajectories (Haber, 2016; Watanabe, 2018, 2022). Thus, the potential role of speciation rates in evolvability may depend on the eccentricity of the variational envelope around the taxa within a clade, the resistance of that envelope to external pressures, and the relative constancy of phenotypic changes at speciation within and among clades. Still little-studied are among-clade intrinsic differences that determine such features at this scale, or their mechanistic underpinnings. Analyses of G-matrices and P-matrices in large phylogenies are sorely needed.
-
(b)
Traits can hitchhike on high speciation rates, proliferating in the clades that generate more species per unit time (see Jablonski, 2017b for references). Thus, any attribute that tends to confer high speciation rates, such as low dispersal ability (see Jablonski, 2008a, and for a recent discussion in birds, Tobias et al., 2020), might promote the proliferation of other traits that happen to covary with it among lineages. Such attributes often have significant phylogenetic signal, carrying over among descendant species (Jablonski, 2008a, 2017b; and note Tucker et al.’s (2019) argument that the strongly unbalanced topologies of published clades are consistent with such heritability at the species level). This hitchhiking aspect of species selection in the broad sense is likely to be widespread (Jablonski, 2017b; Polly et al., 2017), so that the apparent evolvability of a trait, or of a clade, should be analyzed in a framework that takes both direct organismic selection and this indirect, cross-level effect into account: among-clade differences in traversal of morphospace might be a function of higher-level traits that promote or damp speciation, rather than covariation structure of organismic traits and their genetic underpinnings. That is, speciation here is the cause of apparent differences in evolvability, rather than an effect, an idea dating back to the beginnings of the punctuated equilibrium discussion (Gould, 2002) and revisited by Futuyma (2015). Caution is needed, however: the relatively small number of species present at a given time for most genera and many families suggests that apparent differences in evolvability could arise by chance, an effect that has been termed species drift (Levinton et al., 1986:178; Gould, 2002:736; Stanley, 1979 [as “phylogenetic drift”]; Turner, 2015; Chevin, 2016; Jablonski, 2017b). Such scaling effects may be important in comparative analyses, although the general scarcity of sustained macroevolutionary trends, as opposed to the increases or decreases in ranges of trait values that can notoriously mimic trends (Gould, 2002; Jablonski, 2017b), has been taken as evidence against the pervasiveness of species drift (Simpson & Müller, 2012). To the extent that drift occurs, it is worth remembering its phenotypic effects are determined in part by underlying covariation structure (e.g. Arnold et al., 2001), reducing the strict adherence of phenotypic trajectories to random walks at both the population and clade level.
-
(c)
Directionality aside, clades having high speciation rates potentially generate more phenotypic experiments per unit time than low-rate clades. And if high-speciation clades tend to accumulate species, all else being equal this will tend to reduce the clade’s extinction risk and thus extend its duration, giving the clade more time to explore morphospace. However, although high speciation rates may sometimes promote rapid expansion or translation through morphospace, they do not guarantee it. Counter-examples are well-documented, particularly cases where high speciation rates lack commensurate expansions in morphospace (see “non-adaptive radiations,” Skelton, 1993; Rundell & Price, 2009; Czekanski-Moir & Rundell, 2019); even clades showing considerable movement through morphospace via speciation may ricochet within a confined portion of the space, as in the “regular” urchins in Fig. 1.
Further undermining a simple relation between speciation rates and evolvability, high speciation rates are often accompanied by a “macroevolutionary tradeoff” (Jablonski, 2008a, 2017b) in which traits that confer high speciation rates also impose high extinction rates. For example, low dispersal ability might elevate speciation rates relative to high-dispersal clades, but a decrease in dispersal also tends to narrow geographic ranges and so increase extinction risk. This covariation of origination and extinction rates, mediated by intrinsic biotic factors, has long been recognized and much-discussed (e.g., Valentine, 1969, 1990; Gould & Eldredge, 1977 [as “increaser” vs “survivor” clades]; Stanley, 1979, 1990; Van Valen, 1985; Sepkoski, 1998; Marshall, 2017; Knope et al., 2020), but the macroevolutionary implications are still not fully explored. For example, the greater volatility (negative and positive excursions in standing diversity) of high origination/high extinction clades increases extinction risk, and thus gives them less time to expand or shift in morphospace, or diminishes the extinction-buffering effects of species richness, and so further weakens the expected association between origination rate and disparity and thus evolvability. Instead, traits that strongly elevate speciation rates may entail the macroevolutionary tradeoff, and so become an extinction trap—not quite a macroevolutionary version of evolutionary suicide or Darwinian extinction (see Parvinen, 2016), in that increased speciation rate is probably more often an indirect consequence of selection for other properties than a direct target of selection, but with similar consequences. At least some hyperdiverse clades appear to accumulate species far faster than they expand in morphospace, for example rodents, which show a fascinating range of morphologies but are far more prolific taxonomically, e.g. Alhajeri & Steppan, 2018; Nations et al., 2021).
Despite these observations, blanket statements that “diversity and disparity appear to be fundamentally decoupled” (Guillerme, Cooper, et al., 2020; Oyston et al., 2015; and many more) are an oversimplification. The observation is certainly true for a single moment in geologic time, such as the present-day, but the dynamics are more complex. The two currencies can accrue at different rates, and even at different times, as implied by macroevolutionary lags, but when disparity increases it tends to do so via branching events, that is via taxonomic diversification. Many of these decoupling statements cite the Cambrian radiation, a unique event in the history of life, and so poor grounds for generalization. The apparent pervasiveness of macroevolutionary lags, with delayed bursts in form, function, and diversity, suggests a literature bias in inventories of diversity-disparity relationships (e.g. P. Wagner, 2010). Thus, while the wide range of potential relationships between diversity and disparity is crucial for understanding the evolutionary process, there is an important mechanistic association, albeit an imprecise one, and a more nuanced, quantitative approach is needed.
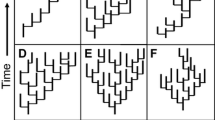
Given the broad range of potential relationships between speciation and a clade’s movement or expansion in morphospace, the clades with the greatest evolvability might be viewed as the ones that disproportionately explore morphospace relative to their speciation rates. Broad morphospace occupation relative to species numbers at a point in time can also be produced by extinction (either random with respect to position in morphospace or against “average” morphologies, see Foote, 1993, 1996), inflating the apparent relationship between diversity and disparity, so that time-series using fossil time-slices in diversity-disparity plots are the most informative approach (Jablonski, 2017b; Wright, 2017). This method has mostly been applied to clades originating under differing conditions (Fig. 3), but comparative analyses of clades responding to the same opportunity, advocated above, would be a valuable extension—revisiting, for example, Eble’s (2000) work comparing holasteroid and spatangoid echinoids, major branches of the irregular clade that differ in their diversity and disparity dynamics, or the contrasting echinoderm clades in the Cambro-Ordovician interval (Deline et al., 2020). Testing potential factors in evolvability, clades having greater modularity or stronger ontogenetic allometry might tend to fall well above the diagonal in Fig. 3, while less modular or more isometric clades below or closer to it.
Evolution in diversity-disparity space. Left: Type 1—Morphology outstrips taxonomic diversification, Type 2—Morphology concordant with taxonomic diversification, Type 3—Morphology trails behind taxonomic diversification. Right, three empirical trajectories, for Cambrian-Ordovician blastozan echinoderms, Jurassic-Cretaceous aporrhaid gastropods, and Ordovician-Carboniferous blastoidean echinoderms. From Jablonski (2017b), which cites sources (Color figure online)
A fuller picture is also needed for how clades move through diversity-disparity space: is an early pulse of phenotypic invention generally balanced by later confinement in morphospace (i.e. shifting from the upper left to lower right in Fig. 3, as implied in many studies, e.g. Cooney et al., 2017, on bird bill evolution)? Alternatively, such a burst might be followed by diffusion in morphospace more proportionate to taxonomic diversification, trending from the upper left in Fig. 3 toward the diagonal: an apparent decline in evolvability but less severe than for clades located in the lower right of the plot. Oyston et al.’s (2015) finding that clades almost always keep acquiring novel traits may suggest trending to the diagonal, but quantitative analyses are needed, and this approach to quantifying diversity-disparity provides an intuitive way to visualize evolvabilty as instances where phenotypic productivity exceeds the stochastic expectation from taxonomic diversification. Further, if ecological opportunity is an important driver interacting with intrinsic evolvability, then combining and partitioning clades within functional categories may yield new insights, as in the underappreciated finding that carnivorous mammals as a functional group show a significant burst in form relative to taxonomic richness after the end-Cretaceous extinction but the constituent clades individually do not (Wesley-Hunt, 2005). Add to this the dependence of early-burst dynamics on taxonomic rank (Foote, 1993; Jablonski, 2017b; Slater & Friscia, 2019), and there is clearly much to do before we approach a robust understanding how evolvability changes over clade history.
Evolvability and Extinction Risk
The macroevolutionary role of evolvability has generally been cast in terms of rates and patterns of origination, and that has been the emphasis here. However, if evolvability entails an ability to respond to changing environments, then lineages with greater evolvability may tend to have lower extinction rates. Such effects could be analyzed at several levels. For example, evolvability might impart higher survival rates to anthropogenically threatened populations (Campbell et al., 2017; Feiner et al., 2021), and to incipient species, e.g. by promoting rapid acquisition of reproductive isolation, or divergence when spatially differentiated populations come back into contact. The latter issue could be especially relevant in clades where ecological speciation is frequent, and the persistence of such populations may be crucial at the inception of diversifications (Gillespie et al., 2020). Paleontologists have long been concerned about, but unable to grapple with, incipient species (e.g. Stanley, 1979): most of these populations are likely to be too ephemeral to enter the fossil record. One approach might be to test a model where isolate formation is primarily an inverse function of dispersal ability, but isolate persistence under sympatry with related species is primarily a positive function of evolvability, using a proxy for evolvability such as phenotypic variation. This two-part model would test whether the canonical stages of speciation are governed by different factors, and bring co-existence of related species more fully into the macroevolutionary arena.
Macroevolutionary “crowding effects” have often been invoked as a basis for large-scale diversity-dependence, but their role at the species level (e.g. Gavina et al., 2018; and see Harvey et al., 2019 more generally on incipient species) has been neglected for long frames despite a large literature on the relation between phylogeny and present-day sympatry. This neglect probably stems in part from the uncertainty imposed by the spatial dynamism of species ranges: geographic overlaps were massively re-arranged during the last deglaciation 11,000 years ago, and evidently have been re-arranged with every glacial-interglacial cycle of the past two Myr (e.g. Jackson & Blois, 2015; Jackson & Williams, 2004; Mottl et al., 2021). Analyses of birds and preliminary analyses of marine bivalves have shown that the groups with the highest diversification rates do not simply carve the world into a mosaic of allopatric species, but also accumulate ecologically similar species in sympatry (secondary or not) (Crouch, 2021). Clades might harbor suites of sympatric taxa for many reasons—an interesting and neglected macroevolutionary issue in its own right—but evolvability can serve as one hypothesis.
At higher levels, not only might clade persistence allow more time to generate new morphologies or functions, but evolvability itself, the ability to generate new morphologies or functions, might promote clade persistence. Broad phenotypic variation, one potential proxy for evolvability, promotes species longevity relative to less variable species in some instances (paleontological examples: Liow, 2007a; Kolbe et al., 2011; neontological examples: González‐Suárez & Revilla, 2013; Forsman & Wennersten, 2016), but counter-examples are known. For example, longer-lived species of Cambrian trilobites tend to have less intraspecific variation but a positive association between geographic range size and species duration, raising the possibility that high rates of morphological evolution might be manifest as variable species of short duration, contrary to the extinction-resistance observed in other studies (Hopkins, 2011). Thus, more work is needed to explore the potential for what Lloyd and Gould (1993) termed “species selection on variability”, where they stress the interaction of that property with the environment to produce “emergent fitness”—lower extinction and/or higher origination rates—at the species level.
Even fewer analyses are available at the clade level. Whether testing for differential clade survivorship across an extinction event, or for clade longevity over many rounds of “normal” extinction, rigorous estimates of phenotypic or functional breadth at these timescales will generally require paleontological data. However, paleo-morphospace analyses tend to examine the effects of extinction on morphospace occupation, rather than vice-versa. Volumes of morphospace occupation are not clearly related to clade persistence in Paleozoic echinoderms (see Deline et al., 2020), and “living fossils” demonstrate the geological persistence of low-diversity, and generally low-disparity, clades (see Lidgard & Love, 2018), both lines of evidence suggesting that extinction risk is not simply an inverse function of extent across morphospace. Knope et al. (2020) do find that functionally diverse clades are more extinction-resistant than functionally homogeneous clades, but phenotypic data are otherwise lacking. If outliers in morphospace tend to be more vulnerable to extinction (e.g. Liow, 2007b; Huang et al., 2015; but see McGowan, 2007), then larger clades may be preferentially trimmed of subclades near the periphery of their overall morphospace, a mechanism for morphospace contraction that may itself raise extinction risk for the larger clade. Further, no analyses have compared extinction risk between clades having anisotropic vs isotropic variation, another potential aspect of evolvability. Again, more comprehensive, comparative analyses are needed.
Temporal and Spatial Patterns: Intrinsic or Extrinsic Factors?
Evolvability does not appear to be constant in time and space. The most frequently cited temporal patterns involve greater evolvability in early metazoan history, and at the inception of clades regardless of their absolute geologic age. Such changes within time series may be best assessed as disparity relative to taxonomic diversity trajectories as in Fig. 3, with high evolvability taken as a disproportionate occupation of morphospace relative to taxonomic richness in a time bin. Such discordance between diversity and disparity suggests that something unusual is going on, and as discussed below, the challenge is to separate intrinsic evolvability from extrinsic opportunities as the primary factor.
Temporal
Debates on the driver(s) of the Cambrian explosion of metazoan form, and its slowdown later in the Paleozoic and to the present day, are essentially asking whether evolvability has changed over time, on a grand scale. The evidence largely supports the view that major clades, and Metazoa overall, underwent a spectacular expansion of morphological and functional breadth in a geologically brief episode (~ 20 Myr according to Paterson et al., 2019) that significantly outpaced taxonomic diversification relative to later events in the history of life (Deline et al., 2020; Erwin & Valentine, 2013; Erwin, 2021b; Jablonski, 2017b). However, mechanisms are still controversial: first, did intrinsic or extrinsic factors drive the rapid expansion in form and function, and second, what then slowed it down? Phylogenetic and paleontological data suggest that many of the developmental tools for building metazoans evolved well before the Cambrian, with a macroevolutionary lag that ended with an extrinsic trigger or opportunity, still not clearly identified (Erwin & Valentine, 2013; Erwin, 2021b). Thus the simple dichotomy between developmental (i.e. intrinsic) and ecological (i.e. extrinsic) mechanisms might be replaced by a “perfect storm” model of mutually reinforcing factors that successively fell into place, none sufficient on its own (Jablonski et al., 2017; Jablonski, 2017b; see Love & Lugar, 2013 for a useful tabulation of hypothesized mechanisms). Increases in gene-regulatory capacity certainly were associated with the Cambrian radiation (reviewed by Erwin, 2021b), but much of that radiation appears to be associated with the redeployment and differentiation of existing developmental pathways. The failure to duplicate the Cambrian burst after the massive end-Permian extinction had been viewed as an argument for a post-Cambrian decline in intrinsic evolvability (but see Foote, 1999), very much in the spirit of the comparative approach suggested here. However, we now know that functional diversity barely dropped after the Permian event despite severe taxonomic losses (Foster & Twitchett, 2014; Edie et al., 2018; and the truly pioneering study by Erwin et al., 1987), suggesting that post-Permian ecological opportunity was not comparable to that of the Cambrian (see Edie et al., 2018; Dunhill et al., 2018 on similar results for other mass extinctions). Most authors currently seem to view the slowdown of the Cambrian explosion in terms of ecological filling of marine habitats.
Comparative studies of variation in Cambrian and post-Cambrian taxa may provide another test for changes in evolvability over the Phanerozoic. Early, low‐dimensionality developmental systems should more fully and evenly occupy morphospace than more derived systems (Borenstein & Krakauer, 2008), and in fact Cambrian trilobite species are notorious for being less sharply defined morphologically than later ones (Webster, 2007). However, we do not yet understand the timescale for “maturation” of developmental systems, and Cambrian biodiversity is extraordinary not just for the presence of apparent intermediates between established clades (Erwin, 2021b notes examples), but for the morphological and functional breadth of Cambrian forms relative to their immediate predecessors, and the rate at which that breadth was achieved. Intraspecific variation is significantly higher in Early Cambrian and more basal trilobites than in later forms (Webster, 2007, 2019), also suggesting a post-Cambrian decrease in evolvability. More work is needed to explore the relevance of intraspecific variation to the dramatic developmental evolution underlying the abrupt rise in Cambrian disparity, and it is unknown whether Cambrian variation is more isotropic than in later times (Jablonski, 2020). Analyses of variation in Cambrian vs later members of more invertebrate clades would be valuable, as would tracking more clades in a standardized diversity-disparity space (Fig. 3, and Wright, 2017; Zhou et al., 2021).
At lower taxonomic levels, evolvability might decline over a clade’s history, regardless of when it originated. This is a long-standing idea, implied for example by Rosa’s Rule that traits vary more in early members of a clade than in later members (Webster, 2019, who notes that the “flexible stem hypothesis” of West-Eberhard (2003) is similar but specifically attributes the early variation to plasticity). Here the macroevolutionary evidence is less clear, if the metric is the growth of disparity relative to diversity––Harmon et al. (2010) detect few early bursts, Hughes et al. (2013) detect many, and Slater and Pennell (2014) attribute Harmon et al.’s result to a lack of statistical power. The work of Hughes et al. is intriguing, but the time of maximum disparity is measured relative to the total duration of each clade, and not against its taxonomic diversity profile through time, and more than half of their included studies show maximum disparity near the middle of a clade’s history. Integrating the early-disparity findings of Hughes et al. (2013) with the macroevolutionary-lag findings of Kröger and Penny (2020), superficially contradictory but actually dealing in different currencies, should clarify matters. Testing for variation in early and late members of clades originating at different points in geologic time would also be valuable—as Webster (2019) observes, the few direct tests have involved trilobite lineages with early members embedded in the Cambrian Explosion, and thus may be addressing a different hypothesis.
If clades do occupy most of their morphospace and functional space early in their histories, several non-exclusive mechanisms might be operating. The rate of production of new character states does seem to slow down in many clades, even when character-state transitions do not (Oyston et al., 2015). However, nearly all clades produce some new character states throughout their history, rather than simply reiterating old states after maximum disparity is reached, by subdividing or migrating through morphospace (Oyston et al., 2015). If these patterns are attributable to intrinsic reductions in evolvability rather than ecological crowding, they imply a relatively weak effect. The apparent tendency for taxonomic diversification to slow with clade age (Henao Diaz et al., 2019) is at least as consistent with crowding effects as with regular, among-clade changes in intrinsic factors such as evolvability.
Perhaps the most provocative evidence for declines in intrinsic evolvability during a clade’s history comes from Wagner’s (2018) analysis of character-state correlations in the fossil record. From a large set of character matrices for fossil taxa, the disparity of clades early in their histories exceeded that expected from two models of independent character change, i.e. continuous change with saturation of a limited space, and elevated early independent rates. Instead, the data were consistent with a model breaking up correlations among characters and forming new ones, arguably analogous to reorganizing the structure of phenotypic variances and covariances—the P-matrix (see Love et al., 2021)—and thus presumably the G-matrix. Developmental data are needed to test this “correlated change‐breakup‐relinkage” model, and a key question is whether these changing linkages unfold across the appropriate timescales, and have the limiting effects on overall phenotypic change that appear to typify the clade histories analyzed by Oyston et al. (2015).
Others suggest that evolvability tends to increase through a clade’s history. Vermeij (2015) argues that younger branches within major animal and plant clades explore a greater portion of morphospace than older ones, explaining this pattern in terms of selection to alleviate energetic tradeoffs. This view implies a ratcheting effect not seen in the analyses by Oyston et al. or Wagner cited above, but those data are at a much finer scale than Vermeij’s examples. Vermeij argues that “versatility’ (which as noted above includes but is not restricted to modularity) has increased overall through time; Goswami et al. (2014) also argue for a net tendency for modularity to increase and integration to decline—implying that the relinkage in P. Wagner’s “correlated change‐breakup‐relinkage” model is more localized within the phenotype than the ancestral state. These are plausible viewpoints that require testing in a common framework. One unexplored possibility is that the increase and later decline of evolvability occurs only at the origin of clades that are founded via an evolutionary novelty sensu G. Wagner (2014), that is a trait lacking homology in the ancestor or that has radically and irreversibly changed from the ancestral state. Testing for declines (or increases!) in apparent evolvability of clades that originated in this way, vs those that more clearly arose in the context of ecological opportunity, may be one way to integrate these rather heterogeneous arguments (Jablonski, 2020). Comparative analysis could also use evolutionary accelerations after mass extinctions to differentiate evolvabilities among contemporaneous clades, to test for differences in expansions in form or function among clades of different ages when encountering the same post-extinction opportunity to test for clade-age effects.
Spatial
Hypotheses for spatial variation in evolvability have long focused on the tropics, stunningly rich in taxonomic diversity and phenotypes, and the fossil record presents an additional, unexpected pattern, with disparity repeatedly emerging in marine invertebrate clades in onshore habitats. Comparing clade dynamics in morphospace across latitudes is challenging in terms of data required and the need to control for the strong latitudinal bias in both paleontological and neontological sampling. Great caution is warranted when maxima in origination rates or standing diversity or disparity are found to lie in the best-sampled regions, usually in the present-day temperate zone, as biases can be so strong that standard methods for factoring them out are ineffective (Valentine et al., 2013). One study that factored out sampling bias in two different ways found a significant tendency for marine invertebrate Orders, as a proxy for significant evolutionary novelty, to originate in the tropics over the past 250 Myr (Jablonski, 1993; Martin et al., 2007). However, data were lacking to test whether higher taxa originated more frequently in the tropics on a per-species basis. Bivalve genera also have preferentially originated in the tropics over the past ~ 15 Myr (Jablonski et al., 2006, 2017), and Kiessling et al. (2010) argued that per-taxon origination rates for marine invertebrate genera were in fact higher in the tropics throughout the Phanerozoic, concentrated in reef habitats. In contrast, phylogenies of extant taxa tend to indicate higher speciation rates in the temperate zone, although here too sampling may be an issue, with many extant tropical species not yet recognized (Freeman & Pennell, 2021). As emphasized here, speciation should be viewed separately from evolvability of form and function.
A far less intuitive pattern occurs along marine depth gradients. Orders of marine invertebrates, again used as proxies for phenotypic novelty, preferentially originated in onshore habitats regularly disturbed by storms or normal wave action (Jablonski & Bottjer, 1990; Jablonski, 2005). This pattern is independent of clade-specific bathymetric diversity gradients, turnover rates, or origination frequencies of constituent genera or within-clade traits, with low-level lineages originating offshore in certain clades and therefore sometimes expanding onshore as well as offshore (see also Jablonski et al., 1997; Tomašových et al., 2014; Bribiesca-Contreras et al., 2017; Franeck & Liow, 2019). The lone morphospace analysis to date is consistent with this finding: two orders of irregular echinoids show greater divergences in disparity at their onshore origins than seen within the clades at any depth once established (Eble, 2000; it would be interesting to plot the branch lengths in Fig. 1 against their bathymetric context). Early vertebrate clades also first appear onshore (Sallan et al., 2018, who unfortunately exaggerate differences with the invertebrate patterns).
As with many other aspects of this overview, we have some provocative patterns, potentially indicating greater evolvability in tropical settings, and in onshore marine environments. New kinds of data and analyses are needed to bring these results more fully into the framework discussed here, and then to address the fundamental question: are they driven by intrinsic factors, as tentatively proposed by Jablonski (2005), or are they promoted by the extrinsic environmental gradients that define them (e.g. Vermeij, 2012)? In other words, do organisms, species, or clades that inhabit warm, shallow settings have properties that enhance evolvability, presumably indirectly selected for by those environments, or do those environments directly promote greater phenotypic change?
Conclusions
Taken together, the data do suggest that intrinsic factors can influence the rate and scope of morphological and functional evolution at large scales. However, major challenges remain in converting these suggestions into a rigorously defined field. Perhaps the central difficulty for macroevolution lies in separating the intrinsic factors from the multitude of potential extrinsic biotic and abiotic drivers in determining vacancies, boundaries, or extents of expansion or transformation in a clade’s morphospace or functional repertoire. When extrinsic factors can be excluded or accounted for, the issue becomes how apparent intrinsic evolvability differences map onto the potential causes of evolvability differences explored here and elsewhere in this volume, and the consequences of those different causes for the persistence or evolutionary lability of clades.
The most powerful analyses will be comparative, with the operational approach advocated here involving tests for among-clade (and perhaps across-time) differences in responses to a shared opportunity, in a macroevolutionary analog to a common-garden experiment. New methods for integrating fossil and present-day data are becoming available, and for macroevolutionary purposes this integration will be essential; one aim of this paper has been to show that there is much raw material and a growing toolkit for moving the field forward. Every among-clade comparison of morphospace occupation or functional diversification is the potential basis for a study of evolvability, particularly when the occupation pattern is informed by phylogeny or explicitly structured over geologic time. We need a more active two-way exchange, predicting macroevolutionary patterns from short-term evolvability estimates, and predicting short-term evolvability and its developmental and genetic underpinnings from macroevolutionary dynamics. Such an exchange should come closer to testing underlying mechanisms and how they play out on the macroevolutionary stage. Evolvability could then become a powerful bridge between micro- and macroevolution. This would not involve simple extrapolation from lower to higher levels, but a way to understand and systematize the many nonlinearities and indirect effects inherent in a multilevel system, as we now understand organic evolution to be.
Data Availability
No new data were generated or analyzed for this review.
References
Aase-Remedios, M. E., & Ferrier, D. E. K. (2021). Improved understanding of the role of gene and genome duplications in chordate evolution with new genome and transcriptome sequences. Frontiers in Ecology and Evolution, 9, 703163.
Abzhanov, A., Kuo, W. P., Hartmann, C., Grant, B. R., Grant, P. R., & Tabin, C. J. (2006). The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature, 442, 563–567.
Alberch, P. (1980). Ontogenesis and morphological diversification. American Zoologist, 20, 653–667.
Alhajeri, B. H., & Steppan, S. J. (2018). Disparity and evolutionary rate do not explain diversity patterns in muroid rodents (Rodentia: Muroidea). Evolutionary Biology, 45, 324–344.
Alix, K., Gérard, P. R., Schwarzacher, T., & Heslop-Harrison, J. S. (2017). Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Annals of Botany, 120, 183–194.
Armbruster, W. S. (2022). Evolvability of plants: Macroevolutionary signals of evolutionary bias and paths of low resistance in floral evolution. In Hansen et al. (2022)
Arnold, S. J., Pfrender, M. E., & Jones, A. G. (2001). The adaptive landscape as a conceptual bridge between micro- and macroevolution. Genetica, 112–113, 9–32.
Audino, J. A., Serb, J. M., & Marian, J. E. A. (2021). Untangling the diversity and evolution of tentacles in scallops, oysters, and their relatives (Bivalvia: Pteriomorphia). Organisms Diversity & Evolution, 21, 145–160.
Barnes, B. D., Sclafani, J. A., & Zaffos, A. (2021). Dead clades walking are a pervasive macroevolutionary pattern. Proceedings of the National Academy of Sciences USA, 118, e201920811.
Benton, M. J. (2009). The Red Queen and the Court Jester: Species diversity and the role of biotic and abiotic factors through time. Science, 323, 728–732.
Betancur-R, R., Ortí, G., & Pyron, R. A. (2015). Fossil-based comparative analyses reveal ancient marine ancestry erased by extinction in ray-finned fishes. Ecology Letters, 18, 441–450.
Beu, A. G., & Raine, J. I. (2009). Revised descriptions of New Zealand Cenozoic Mollusca from Beu and Maxwell (1990). GNS Science Miscellaneous Series 27, online.
Bicknell, R. D., Kimmig, J., Budd, G. E., Legg, D. A., Bader, K. S., Haug, C., Kaiser, D., Laibl, L., Tashman, J. N., & Campione, N. E. (2022). Habitat and developmental constraints drove 330 million years of horseshoe crab evolution. Biological Journal of the Linnean Society. https://doi.org/10.1093/biolinnean/blab173
Boivin, S., Saucède, T., Laffont, R., Steimetz, E., & Neige, P. (2018). Diversification rates indicate an early role of adaptive radiations at the origin of modern echinoid fauna. PLoS ONE, 13, e0194575.
Borenstein, E., & Krakauer, D. C. (2008). An end to endless forms: Epistasis, phenotype distribution bias, and nonuniform evolution. PLoS Computational Biology, 4, e1000202.
Bowers, J. E., & Paterson, A. H. (2021). Chromosome number is key to longevity of polyploid lineages. New Phytologist, 231, 19–28.
Bribiesca-Contreras, G., Verbruggen, H., Hugall, A. F., & O’Hara, T. D. (2017). The importance of offshore origination revealed through ophiuroid phylogenomics. Proceedings of the Royal Society of London, B284, 20170160.
Brombacher, A., Wilson, P. A., Bailey, I., & Ezard, T. H. G. (2017). The breakdown of static and evolutionary allometries during climatic upheaval. American Naturalist, 190, 350–362.
Brown, R. L. (2014). What evolvability really is. British Journal for the Philosophy of Science, 65, 549–572.
Burroughs, R. W. (2019). Phylogenetic and developmental constraints dictate the number of cusps on molars in rodents. Scientific Reports, 9, 10902.
Buss, L. W. (1987). The evolution of individuality. Princeton University Press.
Campbell, C. S., Bean, C. W., & Parsons, K. J. (2017). Conservation evo-devo: Preserving biodiversity by understanding its origins. Trends in Ecology & Evolution, 32, 746–759.
Chapelle, K. E., Benson, R. B., Stiegler, J., Otero, A., Zhao, Q., & Choiniere, J. N. (2020). A quantitative method for inferring locomotory shifts in amniotes during ontogeny, its application to dinosaurs and its bearing on the evolution of posture. Palaeontology, 63, 229–242.
Chevin, L.-M. (2016). Species selection and random drift in macroevolution. Evolution, 70, 513–525.
Clark, J. W., & Donoghue, P. C. J. (2018). Whole-genome duplication and plant macroevolution. Trends in Plant Science, 23, 933–945.
Clark, J. W., Puttick, M. N., & Donoghue, P. C. J. (2019). Origin of horsetails and the role of whole-genome duplication in plant macroevolution. Proceedings of the Royal Society London, B286, 20191662.
Conant, G. C. (2020). The lasting after-effects of an ancient polyploidy on the genomes of teleosts. PLoS One, 15, e0231356.
Conway Morris, S., & Harper, E. (1988). Genome size in conodonts (Chordata): Inferred variations during 270 million years. Science, 241, 1230–1232.
Cooney, C. R., Bright, J. A., Capp, E. J., Chira, A. M., Hughes, E. C., Moody, C. J., Nouri, L. O., Varley, Z. K., & Thomas, G. H. (2017). Mega-evolutionary dynamics of the adaptive radiation of birds. Nature, 542, 344–347.
Cooney, C. R., & Thomas, G. H. (2021). Heterogeneous relationships between rates of speciation and body size evolution across vertebrate clades. Nature Ecology & Evolution, 5, 101–110.
Couzens, A. M., Sears, K. E., & Rücklin, M. (2021). Developmental influence on evolutionary rates and the origin of placental mammal tooth complexity. Proceedings of the National Academy of Sciences USA, 118, e2019294118.
Crouch, N. M. A. (2020). Extinction rates of non-avian dinosaur species are uncorrelated with the rate of evolution of phylogenetically informative characters. Biology Letters, 16, 20200231.
Crouch, N. M. A. (2021). Shared patterns of spatial accumulation of lineages across terrestrial vertebrates. Journal of Biogeography, 48, 1811–1823.
Crouch, N. M. A., & Ricklefs, R. E. (2019). Speciation rate is independent of the rate of evolution of morphological size, shape, and absolute morphological specialization in a large clade of birds. American Naturalist, 193, E78–E91.
Czekanski-Moir, J. E., & Rundell, R. J. (2019). The ecology of nonecological speciation and nonadaptive radiations. Trends in Ecology & Evolution, 34, 400–415.
Davesne, D., Friedman, M., Schmitt, A. D., Fernandez, V., Carnevale, G., Ahlberg, P. E., Sanchez, S., & Benson, R. B. (2021). Fossilized cell structures identify an ancient origin for the teleost whole-genome duplication. Proceedings of the National Academy of Sciences USA, 118, e2101780118.
De Vienne, D. M., Giraud, T., & Gouyon, P. H. (2013). Lineage selection and the maintenance of sex. PLoS ONE, 8, e66906.
Deline, B., Thompson, J. R., Smith, N. S., Zamora, S., Rahman, I. A., Sheffield, S. L., Ausich, W. I., Kammer, T. W., & Sumrall, C. D. (2020). Evolution and development at the origin of a phylum. Current Biology, 30, 1672–1679.
DiFrisco, J., & Jaeger, J. (2021). Homology of process: Developmental dynamics in comparative biology. Interface Focus, 11, 20210007.
Dijkstra, H. H., Warén, A., & Gudmundsson, G. (2009). Pectinoidea (Mollusca: Bivalvia) from Iceland. Marine Biology Research, 5, 207–243.
Donoghue, M. J., & Sanderson, M. J. (2015). Confluence, synnovation, and depauperons in plant diversification. New Phytologist, 207(2), 260–274. https://doi.org/10.1111/nph.13367
Draghi, J., & Wagner, G. P. (2008). Evolution of evolvability in a developmental model. Evolution, 62, 301–315.
Dunhill, A. M., Foster, W. J., Sciberras, J., & Twitchett, R. J. (2018). Impact of the Late Triassic mass extinction on functional diversity and composition of marine ecosystems. Palaeontology, 61, 133–148.
Eble, G. J. (2000). Contrasting evolutionary flexibility in sister groups: Disparity and diversity in Mesozoic atelostomate echinoids. Paleobiology, 26, 56–79.
Edie, S. M., Jablonski, D., & Valentine, J. W. (2018). Contrasting responses of functional diversity to major losses in taxonomic diversity. Proceedings of the National Academy of Sciences USA, 115, 732–737.
Edie, S. M., Khouja, S. C., Collins, K. S., Crouch, N. M. A., & Jablonski, D. (2022). Modularity, integration and disparity in an accretionary skeleton: Analysis of venerid Bivalvia. Proceedings of the Royal Society London B, 289, 20211199.
Erwin, D. H. (2007). Disparity: Morphological pattern and developmental context. Palaeontology, 50, 57–73.
Erwin, D. H. (2021a). A conceptual framework of evolutionary novelty and innovation. Biological Reviews, 96, 1–15.
Erwin, D. H. (2021b). Developmental capacity and the early evolution of animals. Journal of the Geological Society London, 178, jgs020-245.
Erwin, D. H., & Davidson, E. H. (2009). The evolution of hierarchical gene regulatory networks. Nature Reviews Genetics, 10, 141–148.
Erwin, D. H., & Valentine, J. W. (2013). The Cambrian explosion. Ben Roberts.
Erwin, D. H., Valentine, J. W., & Sepkoski, J. J., Jr. (1987). A comparative study of diversification events: The early Paleozoic versus the Mesozoic. Evolution, 41, 1177–1186.
Esquerré, D., Sherratt, E., & Keogh, J. S. (2017). Evolution of extreme ontogenetic allometric diversity and heterochrony in pythons, a clade of giant and dwarf snakes. Evolution, 71, 2829–2844.
Evans, K. M., Larouche, O., Watson, S. J., Farina, S., Habegger, M. L., & Friedman, M. (2021). Integration drives rapid phenotypic evolution in flatfishes. Proceedings of the National Academy of Sciences USA, 118, e2101330118.
Fabre, A. C., Bardua, C., Bon, M., Clavel, J., Felice, R. N., Streicher, J. W., Bonnel, J., Stanley, E. L., Blackburn, D. C., & Goswami, A. (2020). Metamorphosis shapes cranial diversity and rate of evolution in salamanders. Nature Ecology & Evolution, 4, 1129–1140.
Fasanelli, M. N., Carmona, P. M., Soto, I. M., & Tuero, D. T. (2022). Allometry, sexual selection, and evolutionary lines of least resistance shaped the evolution of exaggerated sexual traits within the genus Tyrannus. Journal of Evolutionary Biology. https://doi.org/10.1111/jeb.14000
Feiner, N., Brun-Usan, M., & Uller, Y. (2021). Evolvability and evolutionary rescue. Evolution & Development, 23, 308–319.
Felice, R. N., Randau, M., & Goswami, A. (2018). A fly in a tube: Macroevolutionary expectations for integrated phenotypes. Evolution, 72, 2580–2594.
Felice, R. N., Watanabe, A., Cuff, A. R., Noirault, E., Pol, D., Witmer, L. M., Norell, M. A., O’Connor, P. M., & Goswami, A. (2019). Evolutionary integration and modularity in the archosaur cranium. Integrative and Comparative Biology, 59, 371–382.
Folk, R. A., Stubbs, R. L., Mort, M. E., Cellinese, N., Allen, J. M., Soltis, P. S., Soltis, D. E., & Guralnick, R. P. (2019). Rates of niche and phenotype evolution lag behind diversification in a temperate radiation. Proceedings of the National Academy of Sciences USA, 116, 10874–10882.
Folse, H. J., & Roughgarden, J. (2012). Direct benefits of genetic mosaicism and intraorganismal selection: Modeling coevolution between a long-lived tree and a short-lived herbivore. Evolution, 66, 1091–1113.
Foote, M. (1991). Morphological and taxonomic diversity in a clade’s history: The blastoid record and stochastic simulations. Contributions from the Museum of Paleontology, University of Michigan, 28, 101–140.
Foote, M. (1993). Discordance and concordance between morphological and taxonomic diversity. Paleobiology, 19, 185–204.
Foote, M. (1996). Models of morphological diversification. In D. Jablonski, D. H. Erwin, & J. H. Lipps (Eds.), Evolutionary paleobiology (pp. 62–86). University of Chicago Press.
Foote, M. (1997). The evolution of morphological diversity. Annual Review of Ecology and Systematics, 28, 129–152.
Foote, M. (1999). Morphological diversity in the evolutionary radiation of Paleozoic and post-Paleozoic crinoids. Paleobiology, 25(2), 1–115.
Forsman, A., & Wennersten, L. (2016). Inter-individual variation promotes ecological success of populations and species: Evidence from experimental and comparative studies. Ecography, 39, 630–648.
Foster, W. J., & Twitchett, R. J. (2014). Functional diversity of marine ecosystems after the Late Permian mass extinction event. Nature Geoscience, 7, 233–238.
Franeck, F., & Liow, L. H. (2019). Dissecting the paleocontinental and paleoenvironmental dynamics of the great Ordovician biodiversification. Paleobiology, 45, 221–234.
Freeman, B. G., & Pennell, M. W. (2021). The latitudinal taxonomy gradient. Trends in Ecology & Evolution, 36, 778–786.
Fujita, M. K., Singhal, S., Brunes, T. O., & Maldonado, J. A. (2020). Evolutionary dynamics and consequences of parthenogenesis in vertebrates. Annual Review of Ecology, Evolution, and Systematics, 51, 191–214.
Futuyma, D. J. (2015). Can modern evolutionary theory explain macroevolution? In E. Serrelli & N. Gontier (Eds.), Macroevolution (pp. 29–85). Springer.
Galis, F. (2022). Evolvability of body plans: on phylotypic stages, developmental modularity and an ancient metazoan constraint. In Hansen et al. (2022).
Galis, F., Metz, J. A., & van Alphen, J. J. (2018). Development and evolutionary constraints in animals. Annual Review of Ecology, Evolution, and Systematics, 49, 499–522.
Gavina, M. K. A., Tahara, T., Tainaka, K., Ito, H., Morita, S., Ichinose, G., Okabe, T., Togashi, T., Nagatani, T., & Yoshimura, J. (2018). Multi-species coexistence in Lotka-Volterra competitive systems with crowding effects. Scientific Reports, 8, 1198.
Geiger, M., Evin, A., Sánchez-Villagra, M. R., Gascho, D., Mainini, C., & Zollikofer, C. P. (2017). Neomorphosis and heterochrony of skull shape in dog domestication. Scientific Reports, 7, 13443.
Gerhart, J., & Kirschner, M. (1997). Cells, embryos and evolution: Towards a cellular and developmental understanding of phenotypic variation and evolutionary adaptability. Blackwell.
Gillespie, R. G., Bennett, G. M., De Meester, L., Feder, J. L., Fleischer, R. C., Harmon, L. J., Hendry, A. P., Knope, M. L., Mallet, J., Martin, C., Parent, C. E., Patton, A. H., Pfennig, K. S., Rubinoff, D., Schluter, D., Seehausen, O., Shaw, K. L., Stacy, E., Stervander, M., … Wogan, G. O. U. (2020). Comparing adaptive radiations across space, time, and taxa. Journal of Heredity, 111, 1–20.
Glasauer, S. M., & Neuhauss, S. C. (2014). Whole-genome duplication in teleost fishes and its evolutionary consequences. Molecular Genetics and Genomics, 289, 1045–1060.
González-Suárez, M., & Revilla, E. (2013). Variability in life-history and ecological traits is a buffer against extinction in mammals. Ecology Letters, 16, 242–251.
Gorné, L. D., & Díaz, S. (2019). Meta-analysis shows that rapid phenotypic change in angiosperms in response to environmental change is followed by stasis. American Naturalist, 194, 840–853.
Goswami, A., & Polly, P. D. (2010). The influence of modularity on cranial morphological disparity in Carnivora and Primates (Mammalia). PLoS ONE, 5, e9517.
Goswami, A., Smaers, J. B., Soligo, C., & Polly, P. D. (2014). The macroevolutionary consequences of phenotypic integration: From development to deep time. Philosophical Transactions of the Royal Society London B, 369, 20130254.
Gould, S. J. (1977). Ontogeny and phylogeny. Harvard University Press.
Gould, S. J. (1982). The meaning of punctuated equilibrium and its role in validating a hierarchical approach to macroevolution. In R. Milkman (Ed.), Perspectives on evolution (pp. 183–104). Sinauer.
Gould, S. J. (2002). The structure of evolutionary theory. Belknap Press.
Gould, S. J., & Eldredge, N. (1977). Punctuated equilibria: The tempo and mode of evolution reconsidered. Paleobiology, 3, 115–151.
Guenser, P., Souquet, L., Dolédec, S., Mazza, M., Rigo, M., & Goudemand, N. (2019). Deciphering the roles of environment and development in the evolution of a Late Triassic assemblage of conodont elements. Paleobiology, 45, 440–457.
Guillerme, T., Cooper, N., Brusatte, S. L., Davis, K. E., Jackson, A. L., Gerber, S., Goswami, A., Healy, K., Hopkins, M. J., Jones, M. E., Lloyd, G. T., O’Reilly, J. E., Pate, A., Puttick, M. N., Rayfield, E. J., Saupe, E. E., Sherratt, E., Slater, G. J., Weisbecker, V., … Donoghue, P. C. J. (2020). Disparities in the analysis of morphological disparity. Biology Letters, 16, 20200199.
Guillerme, T., Puttick, M. N., Marcy, A. E., & Weisbecker, V. (2020). Shifting spaces: Which disparity or dissimilarity measurement best summarize occupancy in multidimensional spaces? Ecology and Evolution, 10, 7261–7275.
Haber, A. (2016). Phenotypic covariation and morphological diversification in the ruminant skull. American Naturalist, 187, 576–591.
Hallgrímsson, B., Aponte, J. D., Vidal-García, M., Richbourg, H., Green, R., Young, N. M., Cheverud, J. M., Calof, A., Lander, A., & Marcucio, R. (2022). The generation of continuous and discontinuous variation and the developmental basis for evolvability. In Hansen et al. (2022).
Hallgrímsson, B., Katz, D. C., Aponte, J. D., Larson, J. R., Devine, J., Gonzalez, P. N., Young, N. M., Roseman, C. C., & Marcucio, R. S. (2019). Integration and the developmental genetics of allometry. Integrative and Comparative Biology, 59, 1369–1381.
Halliday, T. J., Dos Reis, M., Tamuri, A. U., Ferguson-Gow, H., Yang, Z., & Goswami, A. (2019). Rapid morphological evolution in placental mammals post-dates the origin of the crown group. Proceedings of the Royal Society London B, 286, 20182418.
Hansen, T. F. (2003). Is modularity necessary for evolvability? Remarks on the relationship between pleiotropy and evolvability. Bio Systems, 69, 83–94.
Hansen, T. F. (2006). The evolution of genetic architecture. Annual Review of Ecology, Evolution, and Systematics, 37, 123–157.
Hansen, T. F. (2011). Epigenetics: Adaptation or contingency? In B. Hallgrimsson & B. K. Hall (Eds.), Epigenetics: Linking genotype and phenotype in development and evolution (pp. 357–376). University of California Press.
Hansen, T. F., Houle, D., Pavlicev, M., & Pélabon, C. (2022). Evolvability: A unifying concept in evolutionary biology? MIT Press.
Hansen, T. F., & Pélabon, C. (2021). Evolvability: A quantitative-genetics perspective. Annual Review of Ecology, Evolution, and Systematics, 52, 153–175.
Harmon, L. J., Losos, J. B., Davies, T. J., Gillespie, R. G., Gittleman, J. L., Jennings, W. B., Kozak, K. H., McPeek, M. A., Moreno-Roark, F., Near, T. J., Purvis, A., Ricklefs, R. E., Schluter, D., Schulte, J. A., II., Seehausen, O., Sidlauskas, B. L., Torres-Carvajal, O., Weir, J. T., & Mooers, A. Ø. (2010). Early bursts of body size and shape evolution are rare in comparative data. Evolution, 64, 2385–2396.
Harper, E. M., & Checa, A. G. (2020). Tightly shut: Flexible valve margins and microstructural asymmetry in pterioid bivalves. Marine Biology, 167, 78.
Harvey, M. G., Singhal, S., & Rabosky, D. L. (2019). Beyond reproductive isolation: Demographic controls on the speciation process. Annual Review of Ecology, Evolution, and Systematics, 50, 75–95.
Hedrick, B. P., Mutumi, G. L., Munteanu, V. D., Sadier, A., Davies, K. T., Rossiter, S. J., Sears, K. E., Dávalos, L. M., & Dumont, E. (2020). Morphological diversification under high integration in a hyper diverse mammal clade. Journal of Mammalian Evolution, 27, 563–575.
Hembry, D. H., & Weber, M. G. (2020). Ecological interactions and macroevolution: A new field with old roots. Annual Review of Ecology, Evolution, and Systematics, 51, 215–243.
Henao Diaz, L. F., Harmon, L. J., Sugawara, M. T., Miller, E. T., & Pennell, M. W. (2019). Macroevolutionary diversification rates show time dependency. Proceedings of the National Academy of Sciences USA, 116, 7403–7408.
Herlitze, I., Marie, B., Marin, F., & Jackson, D. J. (2018). Molecular modularity and asymmetry of the molluscan mantle revealed by a gene expression atlas. GigaScience, 7, giy056.
Hiebert, L. S., Simpson, C., & Tiozzo, S. (2021). Coloniality, clonality, and modularity in animals: The elephant in the room. Journal of Experimental Zoology Part b: Molecular and Developmental Evolution, 336, 198–211.
Holzman, R., Collar, D. C., Mehta, R. S., & Wainwright, P. C. (2011). Functional complexity can mitigate performance trade-offs. American Naturalist, 177, E69–E83.
Hopkins, M. J. (2011). How species longevity, intraspecific morphological variation, and geographic range size are related: A comparison using Late Cambrian trilobites. Evolution, 65, 3253–3273.
Hopkins, M. J., & Smith, A. B. (2015). Dynamic evolutionary change in post-Paleozoic echinoids and the importance of scale when interpreting changes in rates of evolution. Proceedings of the National Academy of Sciences USA, 112, 3758–3763.
Houle, D., & Pélabon, C. (2022). Measuring evolvability. In Hansen et al. (2022).
Houle, D., Bolstad, G. H., van der Linde, K., & Hansen, T. F. (2017). Mutation predicts 40 million years of fly wing evolution. Nature, 548, 447–450.
Huang, S., Roy, K., & Jablonski, D. (2015). Origins, bottlenecks, and present-day diversity: Patterns of morphospace occupation in marine bivalves. Evolution, 69, 735–746.
Hughes, M., Gerber, S., & Wills, M. A. (2013). Clades reach highest morphological disparity early in their evolution. Proceedings of the National Academy of Sciences USA, 110, 13875–13879.
Hunt, G. (2007). Evolutionary divergence in directions of high phenotypic variance in the ostracode genus Poseidonamicus. Evolution, 61, 1560–1576.
Hunt, G., & Yasuhara, M. (2010). A fossil record of developmental events: Variation and evolution in epidermal cell divisions in ostracodes. Evolution & Development, 12, 635–646.
Jablonski, D. (1993). The tropics as a source of evolutionary novelty: The post-Palaeozoic fossil record of marine invertebrates. Nature 364(6433), 142–144. https://doi.org/10.1038/364142a0
Jablonski, D. (2002). Survival without recovery after mass extinctions. Proceedings of the National Academy of Sciences USA, 99, 8139–8144.
Jablonski, D. (2005). Evolutionary innovations in the fossil record: The intersection of ecology, development and macroevolution. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 304B(6), 504–519. https://doi.org/10.1002/jez.b.21075
Jablonski, D. (2008a). Species selection: Theory and data. Annual Review of Ecology, Evolution and Systematics, 39, 501–524.
Jablonski, D. (2008b). Biotic interactions and macroevolution: Extensions and mismatches across scales and levels. Evolution, 62, 715–739.
Jablonski, D. (2017a). Approaches to macroevolution: 1. General concepts and origin of variation. Evolutionary Biology, 44, 427–450.
Jablonski, D. (2017b). Approaches to macroevolution: 2. Sorting of variation, some overarching issues, and general conclusions. Evolutionary Biology, 44, 451–475.
Jablonski, D. (2020). Developmental bias, macroevolution, and the fossil record. Evolution and Development, 22, 103–125.
Jablonski, D., & Bottjer, D. J. (1990). The origin and diversification of major groups: Environmental patterns and macroevolutionary lags. In P. D. Taylor & G. P. Larwood (Eds.), Major evolutionary radiations (pp. 17–57). Clarendon Press.
Jablonski, D., Huang, S., Roy, K., & Valentine, J. W. (2017). Shaping the latitudinal diversity gradient: New perspectives from a synthesis of paleobiology and biogeography. American Naturalist, 189, 1–12.
Jablonski, D., Lidgard, S., & Taylor, P. D. (1997). Comparative ecology of bryozoan radiations: Origin of novelties in cyclostomes and cheilostomes. Palaios, 12, 505–523.
Jablonski, D., Roy, K., & Valentine, J. W. (2006). Out of the Tropics: Evolutionary dynamics of the latitudinal diversity gradient. Science, 314, 102–106.
Jackson, S. T., & Blois, J. L. (2015). Community ecology in a changing environment: Perspectives from the Quaternary. Proceedings of the National Academy of Sciences USA, 112, 4915–4921.
Jackson, S. T., & Williams, J. W. (2004). Modern analogs in Quaternary paleoecology: Here today, gone yesterday, gone tomorrow? Annual Review of Earth and Planetary Sciences, 32, 495–537.
Jernvall, J. (2000). Linking development with generation of novelty in mammalian teeth. Proceedings of the National Academy of Sciences USA, 97, 2641–2645.
Johnson, C. K., & Voss, S. R. (2013). Salamander paedomorphosis: Linking thyroid hormone to life history and life cycle evolution. Current Topics in Developmental Biology, 103, 229–258.
Kiessling, W., Simpson, C., & Foote, M. (2010). Reefs as cradles of evolution and sources of biodiversity in the Phanerozoic. Science, 327, 196–198.
Klingenberg, C. P. (2008). Morphological integration and developmental modularity. Annual Review of Ecology, Evolution, and Systematics, 39, 115–132.
Klingenberg, C. P. (2014). Studying morphological integration and modularity at multiple levels: Concepts and analysis. Philosophical Transactions of the Royal Society of London B, 369, 20130249.
Knope, M. L., Bush, A. M., Frishko, L. O., Heim, N. A., & Payne, J. L. (2020). Ecologically diverse clades dominate the oceans via extinction resistance. Science, 367, 1035–1038.
Kolbe, S. E., Lockwood, R., & Hunt, G. (2011). Does morphological variation buffer against extinction? A test using veneroid bivalves from the Plio-Pleistocene of Florida. Paleobiology, 37, 355–368.
Kraaijeveld, K. (2010). Genome size and species diversification. Evolutionary Biology, 37, 227–233.
Kröger, B., & Penny, A. (2020). Skeletal marine animal biodiversity is built by families with long macroevolutionary lag times. Nature Ecology & Evolution, 4, 1410–1415.
Lamsdell, J. C. (2016). Horseshoe crab phylogeny and independent colonizations of fresh water: Ecological invasion as a driver for morphological innovation. Palaeontology, 59, 181–194.
Lamsdell, J. C. (2021). A new method for quantifying heterochrony in evolutionary lineages. Paleobiology, 47, 363–384.
Lavoie, H., Hogues, H., Mallick, J., Sellam, A., Nantel, A., & Whiteway, M. (2010). Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biology, 8, e1000329.
Le Maître, A., Grunstra, N. D., Pfaff, C., & Mitteroecker, P. (2020). Evolution of the mammalian ear: An evolvability hypothesis. Evolutionary Biology, 47, 187–192.
Lehman, J., & Stanley, K. O. (2013). Evolvability is inevitable: Increasing evolvability without the pressure to adapt. PLoS ONE, 8, e62186.
Lertzman-Lepofsky, G., Mooers, A. Ø., & Greenberg, D. A. (2019). Ecological constraints associated with genome size across salamander lineages. Proceedings of the Royal Society London B, 286, 20191780.
Levin, D. A. (2020). Has the polyploid wave ebbed? Frontiers in Plant Science, 11, 251.
Levinton, J. S., Bandel, K., Charlesworth, B., Müller, G., Nagl, W., Runnegar, B., Selander, R. K., Stearns, S. C., Turner, J. R. G., Urbanek, A. J., & Valentine, J. W. (1986). Organismic evolution: The interaction of microevolutionary and macroevolutionary processes. In D. M. Raup & D. Jablonski (Eds.), Patterns and processes in the history of life (pp. 167–182). Springer.
Levis, N. A., & Pfennig, D. W. (2021). Innovation and diversification via plasticity-led evolution. In D. W. Pfennig (Ed.), Phenotypic plasticity and evolution: Causes, consequences, controversies (pp. 211–240). CRC Press.
Lidgard, S., Carter, M. C., Dick, M. H., Gordon, D. P., & Ostrovsky, A. N. (2012). Division of labor and recurrent evolution of polymorphisms in a group of colonial animals. Evolutionary Ecology, 26, 233–257.
Lidgard, S., & Love, A. C. (2018). Rethinking living fossils. BioScience, 68, 760–770.
Lien, S., Koop, B. F., Sandve, S. R., Miller, J. R., Kent, M. P., Nome, T., Hvidsten, T. R., Leong, J. S., Minkley, D. R., Zimin, A., Grammes, F., Grove, H., Gjuvsland, A., Walenz, B., Hermansen, R. A., von Schalburg, K., Rondeau, E. B., Di Genova, A., Samy, J. K. A., … Davidson, W. S. (2016). The Atlantic salmon genome provides insights into rediploidization. Nature, 533, 200–205.
Liow, L. H. (2007a). Does versatility as measured by geographic range, bathymetric range and morphological variability contribute to taxon longevity? Global Ecology and Biogeography, 16, 117–128.
Liow, L. H. (2007b). Lineages with long durations are old and morphologically average: An analysis using multiple datasets. Evolution, 61, 885–901.
Lister, A. M. (2021). Phenotypic plasticity in the fossil record. In D. W. Pfennig (Ed.), Phenotypic plasticity and evolution: Causes, consequences, controversies (pp. 267–297). CRC Press.
Liu, C., Ren, Y., Li, Z., Hu, Q., Yin, L., Wang, H., Qiao, X., Zhang, Y., Xing, L., Xi, Y., & Jiang, F. (2021). Giant African snail genomes provide insights into molluscan whole-genome duplication and aquatic–terrestrial transition. Molecular Ecology Resources, 21, 478–494.
Lloyd, E. A., & Gould, S. J. (1993). Species selection on variability. Proceedings of the National Academy of Sciences USA, 90, 595–599.
Lomax, B. H., Hilton, J., Bateman, R. M., Upchurch, G. R., Lake, J. A., Leitch, I. J., Cromwell, A., & Knight, C. A. (2014). Reconstructing relative genome size of vascular plants through geological time. New Phytologist, 201, 636–644.
López-Sauceda, J., Malda-Barrera, J., Laguarda-Figueras, A., Solís-Marín, F., & Aragón, J. L. (2014). Influence of modularity and regularity on disparity of Atelostomata sea urchins. Evolutionary Bioinformatics, 10, EBO-S14457.
Losos, J. B. (2010). Adaptive radiation, ecological opportunity, and evolutionary determinism. American Naturalist, 175, 623–639.
Losos, J. B. (2011). Convergence, adaptation, and constraint. Evolution, 65, 1827–1840.
Louca, S., & Pennell, M. W. (2020). Extant timetrees are consistent with a myriad of diversification histories. Nature, 580(7804), 502–505. https://doi.org/10.1038/s41586-020-2176-1
Love, A. C. (2003). Evolvability, dispositions, and intrinsicality. Philosophy of Science, 70, 1015–1027.
Love, A. C., Grabowski, M., Houle, D., Liow, L. H., Porto, A., Tsuboi, M., Voje, K. L., & Hunt, G. (2021). Evolvability in the fossil record. Paleobiology. https://doi.org/10.1017/pab.2021.36
Love, A. C., & Lugar, G. L. (2013). Dimensions of integration in interdisciplinary explanations of the origin of evolutionary novelty. Studies in History and Philosophy of Science Part c: Studies in History and Philosophy of Biological and Biomedical Sciences, 44, 537–550.
Mable, B. K., Alexandrou, M. A., & Taylor, M. I. (2011). Genome duplication in amphibians and fish: An extended synthesis. Journal of Zoology, 284, 151–182.
Machado, F. A., Zahn, T. M. G., & Marroig, G. (2018). Evolution of morphological integration in the skull of Carnivora (Mammalia): Changes in Canidae lead to increased evolutionary potential of facial traits. Evolution, 72, 1399–1419.
Mallarino, R., Grant, P. R., Grant, B. R., Herrel, A., Kuo, W. P., & Abzhanov, A. (2011). Two developmental modules establish 3D beak-shape variation in Darwin’s finches. Proceedings of the National Academy of Sciences USA, 108, 4057–4062.
Mann, A., Pardo, J. D., & Maddin, H. C. (2022). Snake-like limb loss in a carboniferous amniote. Nature Ecology & Evolution, 6, 614–621.
Marek, R. D., Falkingham, P. L., Benson, R. B., Gardiner, J. D., Maddox, T. W., & Bates, K. T. (2021). Evolutionary versatility of the avian neck. Proceedings of the Royal Society London B, 288, 20203150.
Marlow, H. (2018). Evolutionary development of marine larvae. In T. J. Carrier, A. M. Reitzel, & A. Heyland (Eds.), Evolutionary ecology of marine invertebrate larvae (pp. 16–33). Oxford University Press.
Marshall, C. R. (2017). Five palaeobiological laws needed to understand the evolution of the living biota. Nature Ecology & Evolution, 1, 0165.
Martin, C. H., & Richards, E. J. (2019). The paradox behind the pattern of rapid adaptive radiation: How can the speciation process sustain itself through an early burst? Annual Review of Ecology, Evolution, and Systematics, 50, 569–593.
Martin, P.R., Bonier, F., & Tewksbury, J.J. (2007). Revisiting Jablonski (1993): Cladogenesis and range expansion explain latitudinal variation in taxonomic richness. Journal of Evolutionary Biology, 20(3), 930–936. https://doi.org/10.1111/j.1420-9101.2007.01317.x
Martin, R. A. (2019). Scaling of species diversity and body mass in mammals: Cope’s rule and the evolutionary cost of large size. Historical Biology, 31, 1242–1255.
Masterson, J. (1994). Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science, 264, 421–424.
McElwain, J. C., & Steinthorsdottir, M. (2017). Paleoecology, ploidy, paleoatmospheric composition, and developmental biology: A review of the multiple uses of fossil stomata. Plant Physiology, 174, 650–664.
McGhee, G. R., Jr. (2011). Convergent evolution: Limited forms most beautiful. MIT Press.
McGowan, A. J. (2007). Does shape matter? Morphological diversity and differential survivorship among Triassic ammonoid genera. Historical Biology, 19, 157–171.
McMahon, D. P., & Hayward, A. (2016). Why grow up? A perspective on insect strategies to avoid metamorphosis. Ecological Entomology, 41, 505–515.
McNamara, K. J. (2012). Heterochrony: The evolution of development. Evolution: Education and Outreach, 5, 203–218.
Melstrom, K. M., & Irmis, R. B. (2019). Repeated evolution of herbivorous crocodyliforms during the age of dinosaurs. Current Biology, 29, 2389–2395.
Milocco, L., & Salazar-Ciudad, I. (2022). Evolution of the G matrix under nonlinear genotype-phenotype maps. American Naturalist, 199, 420–435.
Mitchell, J. S., Etienne, R. S., & Rabosky, D. L. (2019). Inferring diversification rate variation from phylogenies with fossils. Systematic Biology, 68, 1–18.
Moharrek, F., Taylor, P. D., Silvestro, D., Jenkins, H. L., Gordon, D. P., & Waeschenbach, A. (2022). Diversification dynamics of cheilostome bryozoans based on a Bayesian analysis of the fossil record. Palaeontology, 65, e12586.
Mongiardino Koch, N. (2021). Exploring adaptive landscapes across deep time: A case study using echinoid body size. Evolution, 75, 1567–1581.
Moore, M. P., & Martin, R. A. (2021). Natural selection on adults has trait-dependent consequences for juvenile evolution in dragonflies. American Naturalist, 197, 677–689.
Mottl, O., Flantua, S. G., Bhatta, K. P., Felde, V. A., Giesecke, T., Goring, S., Grimm, E. C., Haberle, S., Hooghiemstra, H., Ivory, S., & Kuneš, P. (2021). Global acceleration in rates of vegetation change over the past 18,000 years. Science, 372, 860–864.
Moulton, D. E., Goriely, A., & Chirat, R. (2020). Mechanics unlocks the morphogenetic puzzle of interlocking bivalved shells. Proceedings of the National Academy of Sciences USA, 117, 43–51.
Müller, J., Scheyer, T. M., Head, J. J., Barrett, P. M., Werneburg, I., Ericson, P. G., Pol, D., & Sánchez-Villagra, M. R. (2010). Homeotic effects, somitogenesis and the evolution of vertebral numbers in recent and fossil amniotes. Proceedings of the National Academy of Sciences USA, 107, 2118–2123.
Murugesan, S. N., Connahs, H., Matsuoka, Y., Das Gupta, M., Tiong, G. J., Huq, M., Gowri, V., Monroe, S., Deem, K. D., Werner, T., & Tomoyasu, Y. (2022). Butterfly eyespots evolved via cooption of an ancestral gene-regulatory network that also patterns antennae, legs, and wings. Proceedings of the National Academy of Sciences USA, 119, e2108661119.
Natale, R., & Slater, G. J. (2022). The effects of foraging ecology and allometry on avian skull shape vary across levels of phylogeny. American Naturalist. https://doi.org/10.1086/720745
Nations, J. A., Mount, G. G., Morere, S. M., Achmadi, A. S., Rowe, K. C., & Esselstyn, J. A. (2021). Locomotory mode transitions alter phenotypic evolution and lineage diversification in an ecologically rich clade of mammals. Evolution, 75, 376–393.
Navalón, G., Marugán-Lobón, J., Bright, J. A., Cooney, C. R., & Rayfield, E. J. (2020). The consequences of craniofacial integration for the adaptive radiations of Darwin’s finches and Hawaiian honeycreepers. Nature Ecology & Evolution, 4, 270–278.
Near, T. J., Dornburg, A., Kuhn, K. L., Eastman, J. T., Pennington, J. N., Patarnello, T., Zane, L., Fernández, D. A., & Jones, C. D. (2012). Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proceedings of the National Academy of Sciences USA, 109, 3434–3439.
Nicol, D. (1958). A survey of inequivalve pelecypods. Journal of the Washington Academy of Sciences, 48, 56–62.
Nijhout, H. F., & McKenna, K. Z. (2017). The origin of novelty through the evolution of scaling relationships. Integrative and Comparative Biology, 57, 1322–1333.
Nunney, L. (1989). The maintenance of sex by group selection. Evolution, 43, 245–257.
Nuño de la Rosa, L. (2017). Computing the extended synthesis: Mapping the dynamics and conceptual structure of the evolvability research front. Journal of Experimental Zoology Part b: Molecular and Developmental Evolution, 328, 395–411.
O’Connor, P. M., Turner, A. H., Groenke, J. R., Felice, R. N., Rogers, R. R., Krause, D. W., & Rahantarisoa, L. J. (2020). Late Cretaceous bird from Madagascar reveals unique development of beaks. Nature, 588, 272–276.
O’Meara, B. C., Smith, S. D., Armbruster, W. S., Harder, L. D., Hardy, C. R., Hileman, L. C., Hufford, L., Litt, A., Magallón, S., Smith, S. A., & Stevens, P. F. (2016). Non-equilibrium dynamics and floral trait interactions shape extant angiosperm diversity. Proceedings of the Royal Society London B, 283, 20152304.
Organ, C. L., Canoville, A., Reisz, R. R., & Laurin, M. (2011). Paleogenomic data suggest mammal-like genome size in the ancestral amniote and derived large genome size in amphibians. Journal of Evolutionary Biology, 24, 372–380.
Organ, C. L., Shedlock, A. M., Meade, A., Pagel, M., & Edwards, S. V. (2007). Origin of avian genome size and structure in nonavian dinosaurs. Nature, 446, 180–184.
Orr, H. A. (2000). Adaptation and the cost of complexity. Evolution, 54, 13–20.
Oyston, J. W., Hughes, M., Gerber, S., & Wills, M. A. (2016). Why should we investigate the morphological disparity of plant clades? Annals of Botany, 117, 859–879.
Oyston, J. W., Hughes, M., Wagner, P. J., Gerber, S., & Wills, M. A. (2015). What limits the morphological disparity of clades? Interface Focus Royal Society of London, 5, 20150042.
Palmer, A. R. (2004). Symmetry breaking and the evolution of development. Science, 306, 828–833.
Panganiban, G., & Rubenstein, J. L. R. (2002). Developmental functions of the Distal-less/Dlx homeobox genes. Development, 129, 4371–4386.
Parsons, K. J., McWhinnie, K., Pilakouta, N., & Walker, L. (2020). Does phenotypic plasticity initiate developmental bias? Evolution and Development, 22, 56–70.
Parvinen, K. (2016). Evolution by natural selection to extinction. Evolutionary Ecology Research, 17, 743–756.
Paterson, J. R., Edgecombe, G. D., & Lee, M. S. (2019). Trilobite evolutionary rates constrain the duration of the Cambrian explosion. Proceedings of the National Academy of Sciences USA, 116, 4394–4399.
Patzkowsky, M. E. (1995). A hierarchical branching model of evolutionary radiations. Paleobiology, 21, 440–460.
Pavlicev, M., & Hansen, T. F. (2011). Genotype-phenotype maps maximizing evolvability: Modularity revisited. Evolutionary Biology, 38, 371–389.
Payne, J. L., & Wagner, A. (2019). The causes of evolvability and their evolution. Nature Reviews Genetics, 20, 24–38.
Pfennig, D. W. (2021). Key questions about phenotypic plasticity. In D. W. Pfennig (Ed.), Phenotypic plasticity and evolution: Causes, consequences, controversies (pp. 55–88). CRC Press.
Piasecka, B., Lichocki, P., Moretti, S., Bergmann, S., & Robinson-Rechavi, M. (2013). The hourglass and the early conservation models—co-existing patterns of developmental constraints in vertebrates. PLoS Genetics, 9, e1003476.
Pie, M. R., & Weitz, J. S. (2005). A null model of morphospace occupation. American Naturalist, 166, E1–E13.
Polly, P. D. (2020). Functional tradeoffs carry phenotypes across the valley of the shadow of death. Integrative and Comparative Biology, 60, 1268–1282.
Polly, P. D., Fuentes-Gonzalez, J., Lawing, A. M., Bormet, A. K., & Dundas, R. G. (2017). Clade sorting has a greater effect than local adaptation on ecometric patterns in Carnivora. Evolutionary Ecology Research, 18, 61–95.
Polly, P. D., & Mock, O. B. (2018). Heritability: The link between development and the microevolution of molar tooth form. Historical Biology, 30, 53–63.
Quental, T. B., & Marshall, C. R. (2010). Diversity dynamics: Molecular phylogenies need the fossil record. Trends in Ecology & Evolution, 25, 434–441.
Rabosky, D. L. (2017). Phylogenetic tests for evolutionary innovation: The problematic link between key innovations and exceptional diversification. Philosophical Transactions of the Royal Society London B, 372, 20160417.
Rabosky, D. L., Santini, F., Eastman, J., Smith, S. A., Sidlauskas, B., Chang, J., & Alfaro, M. E. (2013). Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nature Communications, 4, 1958.
Raff, R. A. (1996). The shape of life: Genes, development, and the evolution of animal form. University of Chicago Press.
Rainford, J. L., Hofreiter, M., Nicholson, D. B., & Mayhew, P. J. (2014). Phylogenetic distribution of extant richness suggests metamorphosis is a key innovation driving diversification in insects. PLoS ONE, 9, e109085.
Ramírez-Barahona, S., Sauquet, H., & Magallón, S. (2020). The delayed and geographically heterogeneous diversification of flowering plant families. Nature Ecology & Evolution, 4, 1232–1238.
Reyes, E., Sauquet, H., & Nadot, S. (2016). Perianth symmetry changes at least 199 times in angiosperm evolution. Taxon, 65, 945–964.
Rhoda, D., Polly, P. D., Raxworthy, C., & Segall, M. (2021). Morphological integration and modularity in the hyperkinetic feeding system of aquatic-foraging snakes. Evolution, 75, 56–72.
Riedl, R. (1978). Order in living organisms. Wiley.
Roemer, E. (1862). Die Kreidebildungen von Texas und ihre Organischen Einschlüsse. A. Marcus.
Rundell, R. J., & Price, T. D. (2009). Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends in Ecology & Evolution, 24, 394–399.
Sallan, L., Friedman, M., Sansom, R. S., Bird, C. M., & Sansom, I. J. (2018). The nearshore cradle of early vertebrate diversification. Science, 362, 460–464.
Saltzberg, C. J., Walker, L. I., Chipps-Walton, L. E., Costa, B. M. A., Spotorno, Á. E., & Steppan, S. J. (2022). Comparative quantitative genetics of the pelvis in four-species of rodents and the conservation of genetic covariance and correlation structure. Evolutionary Biology, 49, 71–83.
Sánchez-Villagra, M. R., Segura, V., Geiger, M., Heck, L., Veitschegger, K., & Flores, D. (2017). On the lack of a universal pattern associated with mammalian domestication: Differences in skull growth trajectories across phylogeny. Royal Society Open Science, 4, 170876.
Saucède, T., Laffont, R., Labruère, C., Jebrane, A., François, E., Eble, G. J., & David, B. (2015). Empirical and theoretical study of atelostomate (Echinoidea, Echinodermata) plate architecture: Using graph analysis to reveal structural constraints. Paleobiology, 41, 436–459.
Schack, C. R., Gordon, D. P., & Ryan, K. G. (2019). Modularity is the mother of invention: A review of polymorphism in bryozoans. Biological Reviews, 94, 773–809.
Schluter, D. (1996). Adaptive radiation along genetic lines of least resistance. Evolution, 50, 1766–1774.
Schoen, D. J., & Schultz, S. T. (2019). Somatic mutation and evolution in plants. Annual Review of Ecology, Evolution, and Systematics, 50, 49–73.
Seilacher, A. (1984). Constructional morphology of bivalves: Evolutionary pathways in primary versus secondary soft-bottom dwellers. Palaeontology, 27, 207–237.
Sepkoski, J. J., Jr. (1998). Rates of speciation in the fossil record. Philosophical Transactions of the Royal Society of London B, 353, 315–326.
Sherratt, E., Alejandrino, A., Kraemer, A. C., Serb, J. M., & Adams, D. C. (2016). Trends in the sand: Directional evolution in the shell shape of recessing scallops (Bivalvia: Pectinidae). Evolution, 70, 2061–2073.
Shi, J. J., Westeen, E. P., & Rabosky, D. L. (2021). A test for rate-coupling of trophic and cranial evolutionary dynamics in New World bats. Evolution, 75, 861–875.
Shubin, N., Tabin, C., & Carroll, S. (2009). Deep homology and the origins of evolutionary novelty. Nature, 457, 818–823.
Sidlauskas, B. L. (2008). Continuous and arrested morphological diversification in sister clades of characiform fishes: A phylomorphospace approach. Evolution, 62, 3135–3156.
Simões, T. R., Vernygora, O., Caldwell, M. W., & Pierce, S. E. (2020). Megaevolutionary dynamics and the timing of evolutionary innovation in reptiles. Nature Communications, 11, 3322.
Simpson, C. (2012). The evolutionary history of division of labour. Proceedings of the Royal Society London B, 279, 116–121.
Simpson, C., Herrera-Cubilla, A., & Jackson, J. B. C. (2020). How colonial animals evolve. Science Advances, 6, eaaw9530.
Simpson, C., & Müller, J. (2012). Species selection in the molecular age. In J. Müller & R. Asher (Eds.), From clone to bone: The synergy of morphological and molecular tools in paleobiology (pp. 116–134). Cambridge University Press.
Simpson, G. G. (1944). Tempo and mode in evolution. Columbia University Press.
Skelton, P. W. (1979). Preserved ligament in a radiolitid rudist bivalve and its implication of mantle marginal feeding in the group. Paleobiology, 5, 90–106.
Skelton, P. W. (1985). Preadaptation and evolutionary innovation in rudist bivalves. Special Papers in Palaeontology, 33, 159–173.
Skelton, P. W. (1993). Adaptive radiation: Definition and diagnostic tests. In D. R. Lees & D. Edwards (Eds.), Evolutionary patterns and processes (pp. 45–58). Academic Press.
Slater, G. J. (2013). Phylogenetic evidence for a shift in the mode of mammalian body size evolution at the Cretaceous-Palaeogene boundary. Methods in Ecology & Evolution, 4, 734–744.
Slater, G. J., & Friscia, A. R. (2019). Hierarchy in adaptive radiation: A case study using the Carnivora (Mammalia). Evolution, 73, 524–539.
Slater, G. J., Harmon, L. J., & Alfaro, M. E. (2012). Integrating fossils with molecular phylogenies improves inference of trait evolution. Evolution, 66, 3931–3944.
Slater, G. J., & Pennell, M. W. (2014). Robust regression and posterior predictive simulation increase power to detect early bursts of trait evolution. Systematic Biology, 63, 293–308.
Smith, A. B. (2001). Probing the cassiduloid origins of clypeasteroid echinoids using stratigraphically restricted parsimony analysis. Paleobiology, 27, 392–404.
Smith, A. B. (2005). Growth and form in echinoids: The evolutionary interplay of plate accretion and plate addition. In D. E. G. Briggs (Ed.), Evolving form and function: Fossils and development (pp. 181–196). Yale Peabody Museum.
Soltis, P. S., Liu, X., Marchant, D. B., Visger, C. J., & Soltis, D. E. (2014). Polyploidy and novelty: Gottlieb’s legacy. Philosophical Transactions of the Royal Society of London B, 369, 20130351.
Stanley, S. M. (1979). Macroevolution. Freeman.
Stanley, S. M. (1990). The general correlation between rate of speciation and rate of extinction: Fortuitous causal linkages. In R. M. Ross & W. D. Allmon (Eds.), Causes of evolution (pp. 103–127). University of Chicago Press.
Stayton, C. T. (2019). Performance in three shell functions predicts the phenotypic distribution of hard-shelled turtles. Evolution, 73, 720–734.
Stayton, C. T., O’Connor, L. F., & Nisivoccia, N. M. (2018). The influence of multiple functional demands on morphological diversification: A test on turtle shells. Evolution, 72(9), 1933–1949. https://doi.org/10.1111/evo.13561
Sterelny, K., & Griffiths, P. E. (1999). Sex and death: An introduction to philosophy of biology. University of Chicago Press.
Streelman, J. T., Peichel, C. L., & Parichy, D. M. (2007). Developmental genetics of adaptation in fishes: The case for novelty. Annual Review of Ecology, Evolution, and Systematics, 38, 655–681.
Stroud, J. T., & Losos, J. B. (2016). Ecological opportunity and adaptive radiation. Annual Review of Ecology, Evolution, and Systematics, 47, 507–532.
Stubbs, T. L., Pierce, S. E., Rayfield, E. J., & Anderson, P. S. (2013). Morphological and biomechanical disparity of crocodile-line archosaurs following the end-Triassic extinction. Proceedings of the Royal Society B, 280, 20131940.
Sumiyama, K., & Tanave, A. (2020). The regulatory landscape of the Dlx gene system in branchial arches: Shared characteristics among Dlx bigene clusters and evolution. Development, Growth & Differentiation, 62, 355–362.
Tajika, A., Nützel, A., & Klug, C. (2018). The old and the new plankton: Ecological replacement of associations of mollusc plankton and giant filter feeders after the Cretaceous? PeerJ, 6, e4219.
Taylor, M. P., & Wedel, M. J. (2013). Why sauropods had long necks; and why giraffes have short necks. PeerJ, 1, e36.
Taylor, P. D., & Waeschenbach, A. (2015). Phylogeny and diversification of bryozoans. Palaeontology, 58, 585–599.
Teichert, S., & Nützel, A. (2015). Early Jurassic anoxia triggered the evolution of the oldest holoplanktonic gastropod Coelodiscus minutus by means of heterochrony. Acta Palaeontologica Polonica, 60, 269–276.
Thomson, K. S., & Muraszko, K. (1978). Estimation of cell size and DNA content in fossil fishes and amphibians. Journal of Experimental Zoology, 205(2), 315–320. https://doi.org/10.1002/jez.1402050216
Thuy, B., Eriksson, M. E., Kutscher, M., Lindgren, J., Numberger-Thuy, L. D., & Wright, D. F. (2022). Miniaturization during a Silurian environmental crisis generated the modern brittle star body plan. Communications Biology, 5, 14.
Tilman, A. R., & Tilman, D. (2020). Evolution, speciation, and the persistence paradox. In A. Dobson, D. Tilman, & R. D. Holt (Eds.), Unsolved problems in ecology (pp. 160–176). Princeton University Press.
Tobias, J. A., Ottenburghs, J., & Pigot, A. L. (2020). Avian diversity: Speciation, macroevolution, and ecological function. Annual Review of Ecology, Evolution, and Systematics, 51, 533–560.
Tomašových, A., Dominici, S., Zuschin, M., & Merle, D. (2014). Onshore–offshore gradient in metacommunity turnover emerges only over macroevolutionary time-scales. Proceedings of the Royal Society of London B, 281, 20141533.
Tomiya, S., & Miller, L. K. (2021). Why aren’t rabbits and hares larger? Evolution, 75, 847–860.
True, J. R., & Haag, E. S. (2001). Developmental system drift and flexibility in evolutionary trajectories. Evolution & Development, 3, 109–119.
Truman, J. W. (2019). The evolution of insect metamorphosis. Current Biology, 29, R1252–R1268.
Tsuboi, M., van der Bijl, W., Kopperud, B. T., Erritzøe, J., Voje, K. L., Kotrschal, A., Yopak, K. E., Collin, S. P., Iwaniuk, A. N., & Kolm, N. (2018). Breakdown of brain–body allometry and the encephalization of birds and mammals. Nature Ecology & Evolution, 2, 1492–1500.
Tucker, C. M., Aze, T., Cadotte, M. W., Cantalapiedra, J. L., Chisholm, C., Díaz, S., Grenyer, R., Huang, D., Mazel, F., Pearse, W. D., & Pennell, M. W. (2019). Assessing the utility of conserving evolutionary history. Biological Reviews, 94, 1740–1760.
Turner, D. (2015). Historical contingency and the explanation of evolutionary trends. In P.-A. Braillard & C. Malaterre (Eds.), Explanation in biology (pp. 73–90). Springer.
Uller, T., Moczek, A. P., Watson, R. A., Brakefield, P. M., & Laland, K. N. (2018). Developmental bias and evolution: A regulatory network perspective. Genetics, 209, 949–966.
Urdy, S., Wilson, L. A. B., Haug, J. T., & Sánchez-Villagra, M. R. (2013). On the unique perspective of paleontology in the study of developmental evolution and biases. Biological Theory, 8, 293–311.
Ushatinskaya, G. T., & Parkhaev, PYu. (2005). Preservation of imprints and casts of cells of the outer mantle epithelium in the shells of Cambrian brachiopods, mollusks, and problematics. Paleontological Journal, 39, 251–263.
Valentine, J. W. (1969). Patterns of taxonomic and ecological structure of the shelf benthos during Phanerozoic time. Palaeontology, 12, 684–709.
Valentine, J. W. (1990). The macroevolution of clade shape. In R. M. Ross & W. D. Allmon (Eds.), Causes of evolution (pp. 128–150). University of Chicago Press.
Valentine, J. W., Jablonski, D., Krug, A. Z., & Berke, S. K. (2013). The sampling and estimation of marine paleodiversity patterns: Implications of a Pliocene model. Paleobiology, 39, 1–20.
Valentine, J. W., & Marshall, C. R. (2015). Fossil and transcriptomic perspectives on the origins and success of metazoan multicellularity. In I. Ruiz-Trillo & A. M. Nedelcu (Eds.), Evolutionary transitions to multicellular life (pp. 31–46). Springer.
Vamosi, J. C., Magallón, S., Mayrose, I., Otto, S. P., & Sauquet, H. (2018). Macroevolutionary patterns of flowering plant speciation and extinction. Annual Review of Plant Biology, 69, 685–706.
Van Valen, L. (1975). Group selection, sex, and fossils. Evolution, 29, 87–94.
Van Valen, L. M. (1985). A theory of origination and extinction. Evolutionary Theory, 7, 133–142.
Van Valkenburgh, B. (2007). Déjà vu: The evolution of feeding morphologies in the Carnivora. Integrative and Comparative Biology, 47, 147–163.
Vermeij, G. J. (1974). Adaptation, versatility, and evolution. Systematic Zoology, 22, 466–477.
Vermeij, G. J. (1987). Evolution and escalation. Princeton University Press.
Vermeij, G. J. (2012). Crucibles of creativity: The geographic origins of tropical molluscan innovations. Evolutionary Ecology, 26, 357–373.
Vermeij, G. J. (2015). Forbidden phenotypes and the limits of evolution. Interface Focus, Royal Society of London, 5, 20150028.
Voje, K. L., Bell, M. A., & Stuart, Y. E. (2022). Evolution of static allometry and constraint on evolutionary allometry in a fossil stickleback. Journal of Evolutionary Biology, 35, 423–438.
Voje, K. L., Hansen, T. F., Egset, C. K., Bolstad, G. H., & Pélabon, C. (2014). Allometric constraints and the evolution of allometry. Evolution, 68, 866–885.
Wagner, G. P. (2022). Models of contingent evolvability suggest dynamical instabilities in body shape evolution. In Hansen et al. (2022).
Wagner, G. P. (2014). Homology, genes, and evolutionary innovation. Princeton University Press.
Wagner, G. P., & Altenberg, L. (1996). Complex adaptations and the evolution of evolvability. Evolution, 50, 967–976.
Wagner, P. J. (2000). Exhaustion of morphologic character states among fossil taxa. Evolution, 54, 365–386.
Wagner, P. J. (2010). Paleontological perspectives on morphological evolution. In D. Futuyma, J. Levinton, M. Bell, & W. Eanes (Eds.), Evolution since Darwin: The first 150 years (pp. 451–478). Sinauer.
Wagner, P. J. (2018). Early bursts of disparity and the reorganization of character integration. Proceedings of the Royal Society of London B, 285, 20181604.
Wake, D. B. (2009). What salamanders have taught us about evolution. Annual Review of Ecology, Evolution, and Systematics, 40, 333–352.
Waller, T. R. (1998). Origin of the molluscan class Bivalvia and a phylogeny of major groups. In P. A. Johnston & J. W. Haggart (Eds.), Bivalves: An eon of evolution (pp. 1–45). University of Calgary Press.
Watanabe, J. (2018). Clade-specific evolutionary diversification along ontogenetic major axes in avian limb skeleton. Evolution, 72, 2632–2652.
Watanabe, J. (2022). Statistics of eigenvalue dispersion indices: Quantifying the magnitude of phenotypic integration. Evolution, 76, 4–28.
Webster, M. (2007). A Cambrian peak in morphological variation within trilobite species. Science, 317, 499–502.
Webster, M. (2019). Morphological homeostasis in the fossil record. Seminars in Cell & Developmental Biology, 88, 91–104.
Wesley-Hunt, G. D. (2005). The morphological diversification of carnivores in North America. Paleobiology, 31, 35–55.
West-Eberhard, M. J. (2003). Developmental plasticity and evolution. Oxford University Press.
Wimsatt, W. C., & Schank, J. C. (2004). Generative entrenchment, modularity and evolvability: When genic selection meets the whole organism. In G. Schlosser & G. Wagner (Eds.), Modularity in evolution and development (pp. 359–394). University of Chicago Press.
Womack, M. C., Metz, M. J., & Hoke, K. L. (2019). Larger genomes linked to slower development and loss of late-developing traits. American Naturalist, 194, 854–864.
Woods, R. J., Barrick, J. E., Cooper, T. F., Shrestha, U., Kauth, M. R., & Lenski, R. E. (2011). Second-order selection for evolvability in a large Escherichia coli population. Science, 331, 1433–1436.
Wright, D. F. (2017). Phenotypic innovation and adaptive constraints in the evolutionary radiation of Palaeozoic crinoids. Scientific Reports, 7, 13745.
Yang, A. S. (2001). Modularity, evolvability, and adaptive radiations: A comparison of the hemi-and holometabolous insects. Evolution & Development, 3, 59–72.
Young, N. M., Wagner, G. P., & Hallgrímsson, B. (2010). Development and the evolvability of human limbs. Proceedings of the National Academy of Sciences USA, 107(8), 3400–3405. https://doi.org/10.1073/pnas.0911856107
Yu, L., Boström, C., Franzenburg, S., Bayer, T., Dagan, T., & Reusch, T. B. (2020). Somatic genetic drift and multilevel selection in a clonal seagrass. Nature Ecology & Evolution, 4, 952–962.
Zaher, H., Apesteguia, S., & Scanferla, C. A. (2009). The anatomy of the Upper Cretaceous snake Najash rionegrina Apesteguía & Zaher, 2006, and the evolution of limblessness in snakes. Zoological Journal of the Linnean Society, 156, 801–826.
Zenil-Ferguson, R., Burleigh, J. G., Freyman, W. A., Igić, B., Mayrose, I., & Goldberg, E. E. (2019). Interaction among ploidy, breeding system and lineage diversification. New Phytologist, 224, 1252–1265.
Zhou, S., Collins, K. S., Edie, S. M., & Jablonski, D. (2021). Disparification before, during, or after diversification? Testing an Early Burst hypothesis for functional diversity in Early Paleozoic bivalves. Geological Society of America Abstracts with Programs, Annual Meeting (Vol. 53, 141–11.
Acknowledgements
This paper had its start in a much shorter chapter in Hansen et al. (2022). I thank the editors for inviting me to participate in that exciting project; I. Brigandt, K.S. Collins, N.M.A. Crouch, M. Distin, S.M. Edie, F. Galis, T.F. Hansen, S. Huang, M. Pavlicev, C. Pélabon, N.H. Shubin, G. Slater, and S. Zhou for discussions and comments, although none should be held responsible for the end result; and NSF, NASA, and the John Simon Guggenheim Foundation for support.
Author information
Authors and Affiliations
Contributions
D.J. wrote the text.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jablonski, D. Evolvability and Macroevolution: Overview and Synthesis. Evol Biol 49, 265–291 (2022). https://doi.org/10.1007/s11692-022-09570-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-022-09570-4