Abstract
In recent years, Alzheimer’s disease (AD) has been a serious threat to human health. Researchers and clinicians alike encounter a significant obstacle when trying to accurately identify and classify AD stages. Several studies have shown that multimodal neuroimaging input can assist in providing valuable insights into the structural and functional changes in the brain related to AD. Machine learning (ML) algorithms can accurately categorize AD phases by identifying patterns and linkages in multimodal neuroimaging data using powerful computational methods. This study aims to assess the contribution of ML methods to the accurate classification of the stages of AD using multimodal neuroimaging data. A systematic search is carried out in IEEE Xplore, Science Direct/Elsevier, ACM DigitalLibrary, and PubMed databases with forward snowballing performed on Google Scholar. The quantitative analysis used 47 studies. The explainable analysis was performed on the classification algorithm and fusion methods used in the selected studies. The pooled sensitivity and specificity, including diagnostic efficiency, were evaluated by conducting a meta-analysis based on a bivariate model with the hierarchical summary receiver operating characteristics (ROC) curve of multimodal neuroimaging data and ML methods in the classification of AD stages. Wilcoxon signed-rank test is further used to statistically compare the accuracy scores of the existing models. With a 95% confidence interval of 78.87–87.71%, the combined sensitivity for separating participants with mild cognitive impairment (MCI) from healthy control (NC) participants was 83.77%; for separating participants with AD from NC, it was 94.60% (90.76%, 96.89%); for separating participants with progressive MCI (pMCI) from stable MCI (sMCI), it was 80.41% (74.73%, 85.06%). With a 95% confidence interval (78.87%, 87.71%), the Pooled sensitivity for distinguishing mild cognitive impairment (MCI) from healthy control (NC) participants was 83.77%, with a 95% confidence interval (90.76%, 96.89%), the Pooled sensitivity for distinguishing AD from NC was 94.60%, likewise (MCI) from healthy control (NC) participants was 83.77% progressive MCI (pMCI) from stable MCI (sMCI) was 80.41% (74.73%, 85.06%), and early MCI (EMCI) from NC was 86.63% (82.43%, 89.95%). Pooled specificity for differentiating MCI from NC was 79.16% (70.97%, 87.71%), AD from NC was 93.49% (91.60%, 94.90%), pMCI from sMCI was 81.44% (76.32%, 85.66%), and EMCI from NC was 85.68% (81.62%, 88.96%). The Wilcoxon signed rank test showed a low P-value across all the classification tasks. Multimodal neuroimaging data with ML is a promising future in classifying the stages of AD but more research is required to increase the validity of its application in clinical practice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
AD is among the most widespread neurological conditions, affecting over 20 million people globally, and is expected to rise further in the next decades (Stefano et al. 2019). It typically starts in middle or old age. AD is typically considered an irreversible disease without a cure. Both the cardinal clinical symptoms and the disease proteins can be used to classify neurodegenerative diseases, and AD is classified as tauopathies based on the protein (Jeromin and Bowser 2017). The abnormal accumulation of tau protein and amyloid beta (Aβ) caused tauopathies. Although, the pathophysiologic knowledge of Alzheimer’s disease derived from existing ideas such as amyloid beta deposition has greatly aided understanding of the disease process. Amyloid βeta may begin to build up in the brain 20 years before the first indication of AD occurs, whereas the accumulation of Tau protein occurs 15 years before the first symptoms of AD appear (Goenka and Tiwari 2021). The use of biomarkers that indicate pathophysiological alterations suggesting the development of AD has contributed significantly to the effort to identify the disease as early as feasible. Researchers are identifying and refining radiological which is not limited to neuroimaging, genetic, CSF multisensory, speech, electroencephalogram (EEG), and blood biomarkers. Simultaneously, clinical trials are evaluating the impact of biomarkers that potentially slow or stop the progression of AD. Some of the neuroimaging modalities such as functional magnetic resonance imaging (fMRI), fluorodeoxyglucose positron emission tomography (FDG-PET), structural Magnetic resonance imaging (sMRI), and Diffusion Tensor imaging (DTI) have revealed the related structural and behavioral alterations in the brain during the illness process.
The pathogenic aspect of AD was shown by sMRI scanning, which is frequently used to assess morphometric alterations in the brain associated with the loss of synapses, neurons, and dendritic de-arborization in AD progression over time (Salvatore et al. 2018; Dubois et al. 2021). However, structural imaging is insufficient to reflect changes preceding protein buildup. Analysis has shown that metabolic alterations occur before atrophy in people at risk for AD and a functional biomarker can be identified before the specific protein profiles connected to advanced AD using FDG-PET (Ou et al. 2019; Veitch et al. 2022; Kim et al. 2022). Considering the progression of AD and cognitive impairment, fMRI techniques can track AD-related brain damage (Hojjati et al. 2017; Ahmadi et al. 2021; Li et al. 2020). DTI gives information on the structure of the brain in the form of Mean Diffusivity (MD), Fractional Anisotropy (FA), and Echo Planar Imaging (EPI) intensities (De and Chowdhury 2021). Additionally, DTI can spot early microstructural changes in AD patients before they manifest as gross anatomical changes, changes that standard MRI typically misses.
Mild but measurable changes in thinking ability are seen in people with Mild Cognitive Impairment (MCI), and MCI patients have a high chance of developing AD (Kang et al. 2020). MCI is a medical disorder with symptoms that differ from those associated with normal aging. Depending on different stages, Progressive mild cognitive impairment (pMCI), stable mild cognitive impairment (sMCI) (Lu et al. 2022) early mild cognitive impairment (EMCI), and late mild cognitive impairment (LMCI) (Rallabandi et al. 2020), are the four categories under which MCI can be classified. A fundamental change in the assessment of biomarkers / cognitive markers to predict the transition from MCI to Alzheimer’s disease is needed.
Deep Learning (DL) approaches have been used to handle AD diagnosis difficulties successfully in recent years by applying them to neuroimaging single modality. Despite efforts to diagnose AD in the early stages with a single modality, the correctness, and dependability of the findings are open to doubt while thinking about the establishment of established standards for precise AD staging along with fewer AD-related physiological markers (Kim et al. 2022). The functional alterations that occur in the brain areas cannot be evaluated by sMRI, and sMRI is inappropriate for capturing alterations before protein synthesis. Although FDG-PET can deliver a more thorough diagnosis of brain metabolic cognitive function but might not be appropriate to identify the early indications of AD before the neuronal loss occurs.
Considering this, efforts to find a biomarker specific to AD using multimodal neuroimaging data to improve the diagnostic performance of a computer-aided diagnostic (CAD) system have been actively ongoing. The regional distribution of white matter hypometabolism (WMH) associated with Aβ burden, glucose hypometabolism, and gray matter volume reduction has also been examined from MRI and PET (Gaubert et al. 2021; Pham et al. 2022). The pairwise similarity measures for multiple modalities such as VBM-MRI or FDG-PET were utilized for AD analysis (Hao et al. 2020). Furthermore, multimodal connections between tau deposition, gray matter atrophy, hypometabolism, and white matter tract declension in atypical AD were investigated from MRI, PET, and DT (Sintini et al. 2018). The selection of complementary features from each modality is a predominant challenge faced by research communities working in multimodal neuroimaging (Sharma and Mandal 2023). Neuroimaging studies of AD identify different brain regions depending on the imaging modality, and several studies of specific symptoms within AD have been highly inconsistent (Banning et al. 2019). Additionally, the heterogeneity of neuroimaging modality has raised the concern of reproducibility crisis of AD analysis with multimodal neuroimaging data owing to this, a subfield within artificial intelligence (AI), ML, is becoming more common in developing the automatic sophisticated model for multimodal data in early detection of AD.
Earlier this decade, many researchers focused on multimodal learning to gather and combine latent representation data from several neuroimaging techniques. A growing number of studies have looked at MRI and PET extract to learn multilevel and multimodal features by transforming the regional brain images into higher-level characteristics that are more compact (Sarraf and Tofighi 1603; Lu et al. 2018; Abdelaziz et al. 2021; Jin et al. 2022). Similarly, with the recent establishment of multimodal fusion, a growing number of studies have proposed image fusion methods for multimodal neuroimaging analysis in AD diagnosis, and their effectiveness is evaluated using machine learning (ML) algorithms as multimodal classifiers (Lazli et al. 2019; Song et al. 2021). The purpose of the fusion is to have a better contrast, fusion quality, and improved model performance (Muzammil et al. 2020). The successful utilization of multimodal image fusion coupled with ML has shown that it improves the diagnosis of AD (Veshki et al. 2022). The motivation for this study is based on the heterogeneity of neuroimaging modalities and the challenge of the selection of complementary features from each modality (Goenka and Tiwari 2022a). The anatomical and functional changes in the brain linked to AD may be better understood thanks to neuroimaging techniques. However, it is still unclear whether single-modality neuroimaging approaches can reliably and accurately diagnose AD.
This study sought to determine whether multimodal neuroimaging fusion coupled with ML is reliable and effective to distinguish individuals with early symptoms of AD from the terminal stage of AD using a systematic review and measure the effectiveness of its classification using a random effect meta-analysis. A comparable meta-analysis was found in the literature search (Sharma and Mandal 2023), but the procedure utilized in this system is based on (Aggarwal et al. 2021) and is addressing the following Research Questions (RQNs):
-
RQN1: What are the main discoveries and methods used to detect AD using multimodal neuroimaging and ML?".
-
RQN2: What are the various fusion techniques utilized in multimodal neuroimaging studies to facilitate classification?
-
RQN3: What is the percentage usage of various fusion techniques?
-
RQN4: What is the diagnostic accuracy of differentiating between various stages of AD?
-
RQN5: What are the significant differences in the performance of multimodal neuroimaging fusion for the classification tasks?
The contributions of this study are as follows:
-
This study provides a systematic review and meta-analysis of the contribution of machine learning (ML) to the accurate classification of the stages of Alzheimer’s Disease (AD) using multimodal neuroimaging data.
-
The study identifies the potential of multimodal neuroimaging data with ML in accurately classifying different stages of AD. The authors conducted an explainable analysis of the classification algorithms and fusion methods used in the selected studies, which can help researchers and practitioners to understand the strengths and limitations of different methods.
-
The study provides pooled estimates of sensitivity and specificity for differentiating between AD and healthy control participants, as well as for differentiating between different stages of Mild Cognitive Impairment (MCI) and early MCI from NC. These estimates can help researchers and practitioners to evaluate the performance of different methods and to compare their results.
-
The study highlights the need for additional research to increase the validity of the application of multimodal neuroimaging data with ML in clinical practice. This can guide future research and development in this field.
Methodology
This section explains the study’s research techniques, including the research questions, the search procedure, the criteria for inclusion and exclusion, and the selection execution. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) report (Moher et al. 2009), was used to conduct and report this systematic review. A systematic review was conducted to locate studies that used multimodal neuroimaging learning or multimodal neuroimaging fusion to categorize AD phases. Only articles published as a full -text English Language articles between January 2016 and August 2022 (included) were chosen. Articles from before 2016 were excluded because of the methodological (deep learning algorithm and multimodal techniques) gap among earlier research and the criteria used to make them hardly comparable.
We carried out a state-of-the-art search adding phrases together using Boolean operators in IEEE Xplore, Science Direct/Elsevier, ACM Digital Library, and PubMed databases. The relevant subject search terms used are Term A: “Multimodal imaging Fusion” OR “Multimodal Learning”, Term B: “Alzheimer Disease”, Term C: “Mild Cognitive Impairment” OR “MCI”, Term D: “deep learning”. Forward snowballing was also performed on Google Scholar to find any relevant articles. The following rule was created by combining these search keywords: Term A AND Term B AND Term C AND Term D. The eligibility criteria were applied after the removal of duplicates to only choose the articles that included (1) classification of MCI (EMCI or LMCI or pMCI, sMCI) to AD, Stages of AD was diagnosed using internationally accepted scores (3) use of multimodal neuroimaging data (4) Imaging fusion techniques, a (5) classification techniques utilizing ML algorithms (6) accuracy, sensitivity, and specificity for quantitative analysis.
After choosing the appropriate number of studies, the specified facts were extracted for each study: (1) authors and year of publication, (2) Stages, (3) imaging fusion techniques, (4) classification methods, (5) Validation methods, (6) performance metrics score.
We also carried out an explainable analysis based on the systematic evaluation conducted on the commonly used XAI algorithms (Jin et al. 2020). These authors focused on Post-hoc XAI algorithms in their evaluation which explained trained black-box models by probing model parameters and categorized Post-hoc XAI into three: Activation-based, Gradient-based, and Perturbation-based. We further classified the fusion methods into abstraction levels and Performance evaluation analysis of image fusion algorithms based on evaluation conducted by Hermessi et al. (2021).
Data synthesis and analysis were carried out using a metadta statistical program that pools diagnostic test data in Stata. The HSROC model is applied to calculate pooled sensitivity and specificity of selected studies. within- and between-study heterogeneity, along with the correlation between sensitivity and specificity, are all taken into consideration by the hierarchical model (Lee et al. 2015). The command "metandi tp fp fn tn" is used to get the diagnostic odds ratio (DOR), pooled sensitivity, pooled specificity, and likelihood ratio (LR). HSROC is achieved by utilizing the command command “metandiplot tp fp fn tn”. Studies with the same type of diagnosis are considered for meta-analysis. Wilcoxon signed-rank test (Derrac et al. 2011), is utilized to statistically compare the accuracy scores of the existing models and determine if there are significant differences in their performance when using multimodal neuroimaging fusion for the classification of pMCI versus sMCI, MCI versus NC, AD versus NC, and EMCI versus NC.
Results
Search and Study selection
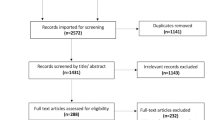
The flow of the survey procedure, as shown in Fig. 1, depicts the analytical review process and the selection of relevant articles at various phases. Database search yielded 2299 results, forward snowballing (Google Scholar) yielded another 50 records, and a total number of 2349 studies were returned from the search. After removing duplicates found due to the combined search, 2247 abstracts were screened. Of these, the 1948 articles did not fulfill the eligibility criteria based on title, abstract, and conclusion. Two hundred and ninety-nine full papers were individually accessed, and 213 Papers were excluded at this stage. 47 papers fulfilled inclusion criteria for the systematic review and contained data for accuracy, sensitivity, and sensitivity for meta-analysis as depicted in Fig. 1, and this expressed the generic answer to RQN1 while the details are provided in subsections of this section.
Summary of the selected studies are represented in Table 1 while Table 2 provides the attributes of the included participants.
Datasets
All studies analyzed in this systematic review used the ADNI dataset, except the study (Schouten et al. 2016) that used the OASIS dataset.
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Marcus et al. 2007) dataset is a publicly available dataset that has been used in many studies on Alzheimer’s Disease (AD). ADNI is a collaborative research effort involving multiple institutions and funded by the National Institutes of Health. The ADNI dataset includes longitudinal data from individuals with AD, Mild Cognitive Impairment (MCI), and healthy control (NC) participants. The data includes clinical assessments, cognitive tests, genetic information, and multimodal neuroimaging data from Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), and cerebrospinal fluid biomarkers. The data typically includes images of brain structures, such as gray matter, white matter, and cerebrospinal fluid, as well as functional images of brain activity, such as regional cerebral blood flow or glucose metabolism.
The Open Access Series of Imaging Studies (OASIS) (Pan et al. 2021b) dataset is a publicly available dataset that contains neuroimaging data and clinical information from individuals with and without dementia. The dataset was created to provide a resource for researchers to study the brain and its changes over time in the context of normal aging and neurodegenerative diseases such as Alzheimer’s disease. The OASIS dataset includes T1-weighted MRI scans, demographic information, and cognitive test scores from over 1,500 individuals. The dataset is divided into two subsets: a cross-sectional dataset and a longitudinal dataset. The cross-sectional dataset includes MRI scans and clinical data from over 400 individuals with Alzheimer’s disease, mild cognitive impairment, and cognitively normal individuals. The longitudinal dataset includes MRI scans and clinical data from over 500 cognitively normal individuals, some of whom went on to develop cognitive impairment or Alzheimer’s disease during the study period. The OASIS dataset has been widely used in research on Alzheimer’s disease and other neurodegenerative diseases, as well as in studies on normal aging and brain development. It has contributed to the development and validation of machine learning models for Alzheimer’s disease diagnosis and classification, as well as to the study of structural changes in the brain over time(Pan et al. 2021b). The OASIS dataset is a valuable resource for researchers studying the brain and its changes over time in the context of aging and neurodegenerative diseases.
The multimodal neuroimaging data used in these datasets provide a rich source of information for machine learning algorithms to identify patterns and classify different stages of AD.
Baseline methods
Baseline methods for AD recognition and stage classification using the ADNI dataset typically involve using clinical and cognitive assessments, as well as neuroimaging data such as Magnetic Resonance Imaging (MRI) and Positron Emission Tomography (PET) scans. In terms of clinical assessments, commonly used measures include the Mini-Mental State Examination (MMSE), Clinical Dementia Rating (CDR), and the Alzheimer’s Disease Assessment Scale-Cognitive subscale (ADAS-Cog). These assessments can help diagnose and stage AD based on the severity of cognitive impairment. Neuroimaging data can also be used for AD recognition and stage classification. MRI scans can be used to measure brain volume, cortical thickness, and hippocampal atrophy, which are all known to be associated with AD. PET scans can be used to measure the accumulation of beta-amyloid and tau proteins, which are also biomarkers of AD. Baseline methods for AD recognition and stage classification using the ADNI dataset typically involve using these clinical and cognitive assessments, as well as neuroimaging data, to identify individuals with AD or mild cognitive impairment, and to differentiate them from healthy control participants. Machine learning algorithms can be applied to these baseline methods to develop more accurate and objective methods for Alzheimer’s disease recognition and stage classification. Several machine learning algorithms can be used as baseline methods for AD recognition and stage classification using the ADNI dataset. Here are some examples:
-
Logistic regression: This is a type of linear model that can be used for binary classification problems (e.g., AD vs. healthy controls). Logistic regression can be used to model the relationship between the input features (e.g., clinical assessments, and neuroimaging data) and the binary outcome variable (e.g., AD vs. healthy controls).
-
Random forest: This is an ensemble learning method that can be used for classification problems. Random forest combines multiple decision trees to make a final prediction. Each tree is trained on a random subset of the input features, and the final prediction is based on the majority vote of all the trees.
-
Support vector machines (SVM): This is a type of linear model that can be used for binary classification problems. SVM finds a hyperplane that separates the input data into two classes (e.g., AD vs. healthy controls). The hyperplane is chosen to maximize the margin between the two classes.
-
Convolutional neural networks (CNN): This is a type of deep learning model that can be used for image analysis tasks, such as MRI or PET scans. CNNs can automatically learn hierarchical representations of the input data and are commonly used for object recognition tasks. In the context of AD recognition, CNNs can be used to identify patterns in neuroimaging data that are indicative of AD or MCI.
These machine-learning algorithms can be used as baseline methods for AD recognition and stage classification and can provide a starting point for developing more accurate and sophisticated models. It is important to note that the choice of algorithm will depend on the specific task and the characteristics of the input data. Metaheuristics approach such as spider monkey optimization algorithm, Cuckoo Search optimization, Bat Inspired Algorithm, Ant Lion Optimization, and Moth Flame Optimization has been hybridized with ML, and utilized in CoVID-19, lung cancer, Retinal artery vein, Chronic Kidney, and diabetes respectively (Kaur et al. 2023). Metaheuristics with ML have also been utilized in the diagnosis of AD in MRI images (Shankar et al. 2019; Chitradevi et al. 2021; Sayed et al. 2017). However, it is important to note that the utilization of metaheuristics in combination with ML techniques has been limited in the context of multimodal neuroimaging.
Features of neuroimaging data
The ranking of features of neuroimaging data from the ADNI and OASIS datasets that have the greatest impact on the medical diagnosis and stage classification of AD may vary depending on the specific machine learning algorithm and dataset used. However, some studies have identified specific features that are consistently important across multiple studies. For example, in a study by Liu et al. (2015) that used the ADNI dataset, the authors found that the most important features for distinguishing AD from healthy controls were gray matter volume in the medial temporal lobe and the entorhinal cortex. In a similar study by Kung et al. (2021) that also used the ADNI dataset, the authors found that cortical thickness in the entorhinal cortex and the inferior temporal gyrus were the most important features for distinguishing AD from healthy controls. In another study by Gu et al. (2022) that used the ADNI dataset, the authors found that the most important features for distinguishing AD from healthy controls were gray matter volume in the hippocampus, amygdala, and temporal lobe, as well as cortical thickness in the medial temporal lobe.
In terms of stage classification, some studies have found that different features may be important for distinguishing between different stages of AD. For example, in a study by Guo et al. (2020) that used the ADNI dataset, the authors found that different features were important for distinguishing between mild cognitive impairment and AD, as compared to distinguishing between mild cognitive impairment and healthy controls. Specifically, cortical thickness in the medial temporal lobe and the inferior temporal gyrus were the most important features for distinguishing mild cognitive impairment from healthy controls, while gray matter volume in the hippocampus, amygdala, and entorhinal cortex were the most important features for distinguishing mild cognitive impairment from AD. The most important features of neuroimaging data for medical diagnosis and stage classification of AD appear to be gray matter volume and cortical thickness in regions of the brain associated with memory and cognitive function, such as the hippocampus, amygdala, and medial temporal lobe. However, the exact features that are most important may vary depending on the specific machine learning algorithm and dataset used, and additional research is needed to further understand the underlying neural mechanisms of AD and how they can be detected using neuroimaging data.
Explainable analysis of the selected studies
The visualization of classification results by the ML models is vital, especially in critical fields like healthcare (Chen et al. 2020). Ensuring that the machine learning model can explain decisions, can also strengthen the possibility to know the model fairness, reliability, and robustness of the model. Explain- ability is also important to debug ML models and make informed decisions about how to improve them. The activation-based method is the most frequently used explanation method for interpreting the predictions of CNN by creating a coarse localization map that highlights the critical areas of the image for the prediction outcome (Selvaraju et al. 2017; Jiang et al. 2021). Gradient-based methods gradient-based visualization methods guided backpropagation, backpropagation, and Grad-CAM are gradient-based visualization methods that determine the gradient of the inference about the input image to retrieve the spatial information of the input called saliency map (Selvaraju et al. 2017; Huff et al. 2021). The perturbation-based method produces a series of perturbed images by modifying the input of the model and observing the changes in the output which are expected to indicate which parts of the input are very important (Ivanovs et al. 2021). The explainable analysis conducted on the forty—seven selected studies is depicted in Table 3
Categorization of image fusion methods into abstraction levels
The answers to RQN2 and RQN3 are provided in this section. The goal of image fusion is to create a merged image by combining information from multiple image (Liu et al. 2018) modalities, and the abstraction level at which information is combined when dealing with complementary information needs to be considered. The fusion methods utilized by the forty- seven studies are classified as three abstraction levels: Pixel-level fusion, Feature-level fusion, and Decision-level fusion (Jin et al. 2020). Pixel–level fusion combined multiple input images which could be captured from different imaging devices or a single type under different parameters settings into a fused image (Liu et al. 2018; Liu et al. 2018; Wang et al. 2023). Feature-level image fusion is an intermediate-level fusion based on the comprehensive analysis of feature information extracted from the information of each image source to form fused information(Wang et al. Feb. 2023; Xiao et al. 2020).Decision-level fusion includes fusion at an advanced level and brings together the interpretations of data from different imaging modalities obtained by local decision-makers based on voting, inference, evidence theory, and fuzzy integral (Xiao et al. 2020; Rajini and Roopa 2017). Figures 2 and 3 gives the percentage usage of each of the fusion level and the classifier for the fused information respectively.
Table 4 summarizes the bivariate and HSROC parameter estimates with their standard errors and approximate 95% confidence interval (CI) in Stata. When differentiating MCI from NC, AD from NC, pMCI from sMCI, and EMCI from NC participants the pooled sensitivity was 83.77% with 95% CI (78.87%, 87.71%), 94.60% with 95% CI (90.76%, 96.89%), 80.41% with 95% CI (74.73%, 85.06%), 86.63 with 95% CI (82.43%, 89.95%), while specificity was 79.16% with 95% CI (70.97%, 87.71%), 93.49 with 95% CI (91.60%, 94.90%), 81.44% with 95% CI (76.32%, 85.66%), 85.68% with 95% CI (81.62%, 88.96%), respectively, as depicted in Table 4.
Figure 4a–d shows the HSROC curve of studies differentiating MCI from NC, AD from NC, pMCI from sMCI, and EMCI from NC participants respectively, and this provides the answer to RQN4. Each study point in Fig. 4 was scaled according to the precision of sensitivity and specificity in the study. The solid circle represents the summary estimate of sensitivity and specificity for each of the diagnosis (MCI vs NC, AD vs NC, pMCI vs sMCI, and EMCI vs NC). The summary point is enclosed by a spotted line denoting the 95% confidence area and a dashed line denoting the 95% prediction area (the area within which one is 95% certain the results of a new study will fall). The pooled DOR for differentiating MCI from NC participants was 19.61% with 95% CI (11.26%, 34.17%), and the pooled DOR for differentiating AD from NC participants was 251.75% with 95% CI (133.30%, 475.44%) while the pooled DOR for differentiating pMCI from sMCI and EMCI from NC participants was 18.01% with 95% CI (11.04%, 29.38%), and 38.80 with 95% CI (22.46%, 67.03%), respectively. Table 5 shows the result of the Wilcoxon signed ranks test for pairwise statistical comparison of the accuracy of the existing model depicted in Table 1 for the classification of pMCI versus sMCI, MCI versus NC, AD versus NC, and EMCI versus NC with 0.90 hypothetical value for comparison. Table 5 provides the answer to RQN5.
Discussion
We looked at research that has already been published on multimodal neuroimaging data with ML algorithms as a fair approach to detecting stages of AD. According to the selected 47 studies in the quantitative analysis, about three studies reported the visualization of feature-level properties using class activation maps. The Post-hoc XAI algorithms for multimodal explanations provided by Jin et al. (2020) are a helpful starting point for the explainable multimodal model. Although most of all the studies included information about the sensitivity and specificity of the model decision the most relevant features to predict AD stages after the fusion of information from different modalities are not analyzed. Only 3 out of the 47 studies presented their results with some visualization of the relevant brain regions for the classification of AD stages. In terms of the fusion methods, we observed that the pixel-level methods (Dwivedi et al. 2022) used techniques based on multiscale decomposition (Wavelet transform), wherein the decomposition transform is used to first break down the source images into multiscale coefficients. Wavelet transforms have been proven to be effective at extracting information details from one image to inject them into another image based on additions, substitutions, or strategy choices. This technique could capture both location and frequency information, and it could extract spatial structures over a range of sizes, thereby being able to separate high frequencies from low frequencies. Most of the studies utilized feature-level methods which operate on features extracted from the images, and the extracted information is achieved using some intelligent computing techniques such as machine learning based methods (Zuo et al. 2021; Xu et al. 2022), region-based algorithms (Pan and Wang 2022), and similarity-matching to content (Dong et al. 2022). Machine learning-based methods (CNN) of multimodality fusion is an effective medical image analysis method (Mathotaarachchi et al. 2017; Huang et al. 2019; Jiang et al. 2021; Liu et al. 2018) for multi-class classification (Goenka and Tiwari 2022b, c). Authors in Daneshtalab et al. (2019) produced an accuracy of 94.2% which is a better performance than that (Qiu et al. 2018) with an accuracy of 84.0%. Both studies fused information extracted from sMRI and DTI images, but the study with machine learning-based methods (Kang et al. 2020) performed better. From Fig. 3, it is shown that feature-level fusion has 75% usage by the included studies.
A preferred abstraction by most of the researchers was feature-level fusion due to its capability of proving more valid results in the case of compatible features (Daneshtalab et al. 2019; Agarwal and Desai 2021). However, the concatenation of compatible features may produce an extremely feature vector that makes the computational load more difficult (Nachappa et al. Apr. 2018). Several other studies used decision-level fusion in which features are ascertained and extracted from each source image, then categorized with regional classifiers, and then decision rules are utilized to combine the information (Peng et al. 2019; Fang et al. 2020). Although decision-level-fusion aimed to support accepted interpretations and comprehensions, the limitation resides in the requirement for prior knowledge-making algorithms to be very complex (Jin et al. 2020; Lahat et al. 2015). Therefore, considering the effect of fusion strategy on the performance of the classification model, we cannot say categorically that a particular fusion strategy is preferable over the others. In all of the fusion levels identified in the included studies, the objective evaluations of the fusion methods’ performances were not considered in all of the included studies, and this evaluation would have helped to assess the image noise, resolution differences between images, and computational complexity from fused images (Kaur et al. 2021). These evaluations would have also provided more insights into the studies utilizing fused images given the percentage of information retained from source images, level of synthetic information produced, and the level of noise (Huang et al. 2020). However, significant progress has been recorded in other domain using pixel fusion-level (Singh et al. 2018; Liu et al. 2021a). Another important finding from this study is that generalization and stability ability of the multimodal model was not further verified as shown in Table 1. None of the studies tested their model in different datasets, and as for validation methods, 29 studies selected cross-validation with different number of folds. Leave -one-out cross validation was selected in 3 studies while random -split validation method was selected in 5 studies. Finally, 10 studies utilized the train/test method of validation.
The results of the meta-analysis are listed in Table 3. We used a bivariate model to directly provide pooled sensitivity and pooled specificity with corresponding 95% CI for four different diagnoses of AD on multimodal neuroimaging data. Sensitivity and specificity are chosen as the main outcome measures in the meta-analysis of diagnostic accuracy studies producing dichotomous index test results because most primary studies report results in pairs of sensitivity and specificity. To the best of our knowledge, this is the first comprehensive review and meta-analysis to look at the diagnostic value of multimodal neuroimaging data for AD diagnosis.
Because the analysis is bivariate, we may test for variations in either sensitivity or specificity or both, between the four diagnoses of AD extracted from the 47 studies. Considering the 47 studies included in the quantitative analysis, the pooled sensitivity and pooled specificity results show that the pooled sensitivity and specificity of studies diagnosing pMCI versus sMCI is significantly lower than that of other studies. It shows that studies with MCI vs NC are a more sensitive test than pMCI versus sMCI, but at the cost of more false positive test findings and a resulting poorer specificity. These results, therefore, suggest favorable sensitivity and specificity of multimodal neuroimaging-based models when compared to single modal neuroimaging–based models. The result of the pooled DOR also indicates heterogeneity between studies, with wide CI indicating the need for more and better-powered studies. The result of the Wilcoxon Signed Ranks Test shows that the obtained P-value from each of the classification tasks is less than the typical significance level of 0.05, which suggests that there is strong evidence to reject the null hypothesis. This indicates that there is a significant difference in the accuracy of the existing models. The results in Table 5 suggest that the classification model can distinguish between individuals with cognitive impairment and those without it, with high accuracy. This finding implies that the classification model is particularly effective at distinguishing between these two groups. The low p-value suggests that the model’s accuracy in classifying individuals as MCI or NC is significantly better than the other three classification tasks.
No study included in the analysis had more than 700 individuals, which raises questions regarding overfitting, most especially for the feature-level fusion. Generally, unsupervised, semi-supervised, supervised, and reinforcement learning are the several subtypes of ML (Kang and Jameson 2018). Most of the studies used supervised algorithms as depicted in Figure with the most common choice being SVM. However, supervised learning is subject to overtraining and overfitting (Kernbach and Staartjes 2022). Thus, the supervised learning algorithm must therefore be continually retrained to retain a good classification performance when exposed to new input data. Also, while semi-supervised learning can infer new knowledge, supervised learning cannot. The former is of higher importance, given the complexity of AD stages. Due to this, cutting–edge semi-supervised learning such as auto-encoder displayed similar performance to supervised ones such as SVM and CNN. Studies utilizing semi-supervised learning approaches such as stacked auto-encoder (Lao and Zhang 2022) or RNN (Feng et al. 2019) reported accuracy, sensitivities, and specificities over 92%, and 83% for AD vs NC, pMCI versus sMCI binary classification respectively, but utilize limited sample sizes as depicted in Table 2. Consequently, there is a need for research into semi-supervised algorithms for categorizing AD stages.
Although the utilization of metaheuristics with ML has shown promise in various medical domains, including the diagnosis of diseases such as COVID-19, lung cancer, retinal artery veins, chronic kidney, and diabetes, its application in multimodal neuroimaging is relatively limited.
Comparison with existing studies
There are a few reviews in this research area. Sharma et al. (2023), conducted a multimodal neuroimaging data review that focused on feature selection, feature scaling, and feature fusion. The conclusion for the further study recommended a robust multimodal ML-based classification model trained on features extracted from an in-house created dataset. Nitika and Shamik (Goenka and Tiwari 2022a) focused on brain-imaging biomarkers based on deep learning frameworks. This review to the best of our knowledge gives a detailed overview of research trends in multimodal neuroimaging for AD and analyses them in various strategies namely: fusion level abstraction, ML method, explainability method, and dataset. This survey followed the procedure laid down in Aggarwal et al. (2021) whose focus was basically on the diagnostic accuracy of ML in medical imaging. Table 6 shows the comparison of this survey with existing ones.
Conclusion
This study shows the potential of multimodal neuroimaging data with machine learning algorithms in accurately classifying different stages of Alzheimer’s Disease. The study performed a systematic review and meta-analysis to evaluate the impact of ML methods on the classification of AD stages. The results show that Machine learning with multimodal neuroimaging data holds great promise for accurately classifying Alzheimer’s disease stages. The study also analyzed the classification algorithms and fusion methods used in the selected studies, providing insights into their strengths and limitations. This information can facilitate researchers in comprehending the diverse methodologies at their disposal and enable them to make judicious choices while devising classification models for Alzheimer’s disease stages, utilizing multimodal neuroimaging. This study also provides the explainability analysis across the selected studies, and it shows that explainability was not available for the majority of the studies, which raises a concern about the reliability of model decisions.
The significant degree of variability or heterogeneity among the research included in the analysis is one of the study’s limitations. This implies that the imaging modalities employed, the image preprocessing methods used, and the classification algorithms used to evaluate the data varied amongst the researchers. Additionally, this review excluded studies that did not report sensitivity and specificity as performance metrics for the classification models. Overall, while the study provides important insights into the potential of machine learning and neuroimaging data for diagnosing AD, these limitations suggest that more research is needed to fully explore and validate these approaches.
Future research should focus on the exploration of other Alzheimer’s disease diagnosis methods with multimodal imaging based on machine learning and metaheuristics approach (Sun et al. 2022; Liu et al. 2021b). The research focus could also be on increasing the sample size for analysis.
References
Abdelaziz M, Wang T, Elazab A (2021) Alzheimer’s disease diagnosis framework from incomplete multimodal data using convolutional neural networks. J Biomed Inform 121:103863. https://doi.org/10.1016/j.jbi.2021.103863
Aderghal K, Khvostikov A, Krylov A, Benois-Pineau J, Afdel K, and Catheline G (2018) Classification of Alzheimer disease on imaging modalities with deep CNNs using cross-modal transfer learning, in 2018 IEEE 31st international symposium on computer-based medical systems (CBMS), pp. 345–350. https://doi.org/10.1109/CBMS.2018.00067.
Agarwal D and Desai S (2021) Multimodal techniques for emotion recognition, in 2021 international conference on computational intelligence and computing applications (ICCICA), pp. 1–6. https://doi.org/10.1109/ICCICA52458.2021.9697294.
Aggarwal R et al (2021) Diagnostic accuracy of deep learning in medical imaging: a systematic review and meta-analysis, npj Digit. Med 4(1):65. https://doi.org/10.1038/s41746-021-00438-z
Ahmadi H, Fatemizadeh E, Motie-Nasrabadi A (2021) Deep sparse graph functional connectivity analysis in AD patients using fMRI data. Comput Methods Programs Biomed 201:105954. https://doi.org/10.1016/j.cmpb.2021.105954
Ahmed OB, Benois-Pineau J, Allard M, Catheline G, Amar CB (2017) Recognition of Alzheimer’s disease and Mild Cognitive Impairment with multimodal image-derived biomarkers and multiple kernel learning. Neurocomputing 220:98–110. https://doi.org/10.1016/j.neucom.2016.08.041
Banning LCP, Ramakers IHGB, Deckers K, Verhey FRJ, Aalten P (2019) Affective symptoms and AT(N) biomarkers in mild cognitive impairment and Alzheimer’s disease: a systematic literature review. Neurosci Biobehav Rev 107:346–359. https://doi.org/10.1016/j.neubiorev.2019.09.014
Chen P, Dong W, Wang J, Lu X, Kaymak U, Huang Z (2020) Interpretable clinical prediction via attention-based neural network. BMC Med Inform Decis Mak 20(3):131. https://doi.org/10.1186/s12911-020-1110-7
Cheng D and Liu M (2017) CNNs based multi-modality classification for AD diagnosis, in 2017 10th international congress on image and signal processing, biomedical engineering and informatics (CISP-BMEI), https://doi.org/10.1109/CISP-BMEI.2017.8302281.
Chitradevi D, Prabha S, Alex P (2021) Diagnosis of Alzheimer disease in MR brain images using optimization techniques. Neural Comput Appl 33(1):223–237. https://doi.org/10.1007/s00521-020-04984-7
Choi H, Jin KH (2018) Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav Brain Res 344:103–109. https://doi.org/10.1016/j.bbr.2018.02.017
Daneshtalab S, Rastiveis H, Hosseiny B (2019) CNN-based feature-level fusion of very high resolution aerial imagery and lidar data. Int Arch Photogramm Remote Sens Spatial Inf Sci XLII-4/W18:279–284. https://doi.org/10.5194/isprs-archives-XLII-4-W18-279-2019
De Stefano C, Fontanella F, Impedovo D, Pirlo G, Scotto di Freca A (2019) Handwriting analysis to support neurodegenerative diseases diagnosis: a review. Pattern Recogn Lett 121:37–45. https://doi.org/10.1016/j.patrec.2018.05.013
De A, Chowdhury AS (2021) DTI based Alzheimer’s disease classification with rank modulated fusion of CNNs and random forest. Expert Syst Appl 169:114338. https://doi.org/10.1016/j.eswa.2020.114338
Derrac J, García S, Molina D, Herrera F (2011) A practical tutorial on the use of nonparametric statistical tests as a methodology for comparing evolutionary and swarm intelligence algorithms. Swarm Evol Comput 1(1):3–18. https://doi.org/10.1016/j.swevo.2011.02.002
Dong A, Zhang G, Liu J (2022) Latent feature representation learning for Alzheimer’s disease classification. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2022.106116
Dubois J, Alison M, Counsell SJ, Hertz-Pannier L, Hüppi PS, Benders MJNL (2021) MRI of the neonatal brain: a review of methodological challenges and neuroscientific advances. J Magn Reson Imaging 53(5):1318–1343. https://doi.org/10.1002/jmri.27192
Dwivedi S, Goel T, Tanveer M, Murugan R, Sharma R (2022) Multimodal fusion-based deep learning network for effective diagnosis of Alzheimer’s disease. IEEE Multimedia 29(2):45–55. https://doi.org/10.1109/MMUL.2022.3156471
Ezzati A et al (2019) Optimizing machine learning methods to improve predictive models of Alzheimer’s disease. J Alzheimer’s Dis 71(3):1027–1036. https://doi.org/10.3233/JAD-190262
Fang X, Liu Z, Xu M (2020) Ensemble of deep convolutional neural networks based multi-modality images for Alzheimer’s disease diagnosis. IET Image Proc 14(2):318–326. https://doi.org/10.1049/iet-ipr.2019.0617
Feng C et al (2019) Deep learning framework for Alzheimer’s disease diagnosis via 3D-CNN and FSBi-LSTM. IEEE Access 7:63605–63618. https://doi.org/10.1109/ACCESS.2019.2913847
Feng C, Elazab A, Yang P, Wang T, Lei B, and Xiao X (2018) 3D convolutional neural network and stacked bidirectional recurrent neural network for Alzheimer’s disease diagnosis, in PRedictive intelligence in MEdicine, Cham, pp. 138–146. https://doi.org/10.1007/978-3-030-00320-3_17.
Forouzannezhad P, Abbaspour A, Li C, Cabrerizo M, and Adjouadi M (2018) A deep neural network approach for early diagnosis of mild cognitive impairment using multiple features, in 2018 17th IEEE international conference on machine learning and applications (ICMLA), pp. 1341–1346. https://doi.org/10.1109/ICMLA.2018.00218.
Gaubert M et al (2021) Topographic patterns of white matter hyperintensities are associated with multimodal neuroimaging biomarkers of Alzheimer’s disease. Alz Res Therapy 13(1):29. https://doi.org/10.1186/s13195-020-00759-3
Goenka N, and Tiwari S (2022a) Alzheimer’s detection using various feature extraction approaches using a multimodal multi‐class deep learning model. In International journal of imaging systems and technology (Vol. 33, Issue 2, pp. 588–609). Wiley. https://doi.org/10.1002/ima.22813
Goenka N and Tiwari S. (2022c) AlzVNet: a volumetric convolutional neural network for multiclass classification of Alzheimer’s disease through multiple neuroimaging computational approaches. In Biomedical signal processing and control (Vol. 74, p. 103500). Elsevier BV. https://doi.org/10.1016/j.bspc.2022.103500
Goenka N, Tiwari S (2022b) Multi-class classification of Alzheimer’s disease through distinct neuroimaging computational approaches using Florbetapir PET scans. Evol Syst. https://doi.org/10.1007/s12530-022-09467-9
Goenka N, Tiwari S (2021) Deep learning for Alzheimer prediction using brain biomarkers. Artif Intell Rev 54(7):4827–4871. https://doi.org/10.1007/s10462-021-10016-0
Gu F, Ma S, Wang X, Zhao J, Yu Y, Song X (2022) Evaluation of feature selection for Alzheimer’s disease diagnosis. Front Aging Neurosci. https://doi.org/10.3389/fnagi.2022.924113
Guo S, Xiao B, Wu C, Alzheimer’s Disease Neuroimaging Initiative (2020) Identifying subtypes of mild cognitive impairment from healthy aging based on multiple cortical features combined with volumetric measurements of the hippocampal subfields. Quant Imaging Med Surg 10(7):1477–1489. https://doi.org/10.21037/qims-19-872
Gupta Y, Kim J-I, Kim BC, Kwon G-R (2020) Classification and graphical analysis of Alzheimer’s disease and its prodromal stage using multimodal features from structural, diffusion, and functional neuroimaging data and the APOE genotype. Front Aging Neurosci 12:238. https://doi.org/10.3389/fnagi.2020.00238
Hao X et al (2020) Multi-modal neuroimaging feature selection with consistent metric constraint for diagnosis of Alzheimer’s disease. Med Image Anal 60:101625. https://doi.org/10.1016/j.media.2019.101625
Hermessi H, Mourali O, Zagrouba E (2021) Multimodal medical image fusion review: theoretical background and recent advances. Signal Process 183:108036. https://doi.org/10.1016/j.sigpro.2021.108036
Hojjati SH, Ebrahimzadeh A, Khazaee A, Babajani-Feremi A (2017) Predicting conversion from MCI to AD using resting-state fMRI, graph theoretical approach and SVM. J Neurosci Methods 282:69–80. https://doi.org/10.1016/j.jneumeth.2017.03.006
Huang Y, Xu J, Zhou Y, Tong T, Zhuang X (2019) and the Alzheimer’s disease neuroimaging initiative (ADNI), Diagnosis of Alzheimer’s disease via multi-modality 3D convolutional neural network. Front Neurosci 13:3. https://doi.org/10.3389/fnins.2019.00509
Huang B, Yang F, Yin M, Mo X, Zhong C (2020) A review of multimodal medical image fusion techniques. Comput Math Methods Med 2020:1–16. https://doi.org/10.1155/2020/8279342
Huff DT, Weisman AJ, Jeraj R (2021) Interpretation and visualization techniques for deep learning models in medical imaging. Phys Med Biol 66(4):04TR01. https://doi.org/10.1088/1361-6560/abcd17
Ivanovs M, Kadikis R, Ozols K (2021) Perturbation-based methods for explaining deep neural networks: a survey. Pattern Recogn Lett 150:228–234. https://doi.org/10.1016/j.patrec.2021.06.030
Jeromin A, Bowser R (2017) Biomarkers in neurodegenerative diseases. Adv Neurobiol 15:491–528. https://doi.org/10.1007/978-3-319-57193-5_20
Jia H, Lao H (2022) Deep learning and multimodal feature fusion for the aided diagnosis of Alzheimer’s disease. Neural Comput Appl 34(22):19585–19598. https://doi.org/10.1007/s00521-022-07501-0
Jiang P-T, Zhang C-B, Hou Q, Cheng M-M, Wei Y (2021) LayerCAM: exploring hierarchical class activation maps for localization. IEEE Trans Image Process 30:5875–5888. https://doi.org/10.1109/TIP.2021.3089943
Jin D et al (2020) Generalizable, reproducible, and neuroscientifically interpretable imaging biomarkers for Alzheimer’s disease. Adv Sci 7(14):2000675. https://doi.org/10.1002/advs.202000675
Jin L, Zhao K, Zhao Y, Che T, Li S (2022) A hybrid deep learning method for early and late mild cognitive impairment diagnosis with incomplete multimodal data. Front Neuroinform 16:843566. https://doi.org/10.3389/fninf.2022.843566
Kang L, Jiang J, Huang J, Zhang T (2020) Identifying early mild cognitive impairment by multi-modality MRI-based deep learning. Front Aging Neurosci 12:206. https://doi.org/10.3389/fnagi.2020.00206
Kang M and Jameson NJ (2018) Machine learning: fundamentals, in Prognostics and health management of electronics, John Wiley & Sons, Ltd, pp. 85–109. https://doi.org/10.1002/9781119515326.ch4.
Kaur H, Koundal D, Kadyan V (2021) Image fusion techniques: a survey. Arch Computat Methods Eng 28(7):4425–4447. https://doi.org/10.1007/s11831-021-09540-7
Kaur S, Kumar Y, Koul A, Kumar Kamboj S (2023) A systematic review on metaheuristic optimization techniques for feature selections in disease diagnosis: open issues and challenges. Arch Comput Methods Eng 30(3):1863–1895. https://doi.org/10.1007/s11831-022-09853-1
Kernbach JM and Staartjes VE (2022) Foundations of machine learning-based clinical prediction modeling: part II–generalization and overfitting, in Machine learning in clinical neuroscience, Cham, 15–21. https://doi.org/10.1007/978-3-030-85292-4_3.
Khvostikov A, Aderghal K, Benois-Pineau J, Krylov A, and Catheline G (2018) 3D CNN-based classification using sMRI and MD-DTI images for Alzheimer disease studies. arXiv:1801.05968 [cs], Accessed: Jan. 04, 2022. [Online]. Available: http://arxiv.org/abs/1801.05968
Kim J, Lee B (2018) Identification of Alzheimer’s disease and mild cognitive impairment using multimodal sparse hierarchical extreme learning machine. Hum Brain Mapp 39(9):3728–3741. https://doi.org/10.1002/hbm.24207
Kim J, Jeong M, Stiles WR, Choi HS (2022) Neuroimaging modalities in Alzheimer’s disease: diagnosis and clinical features. Int J Mol Sci 23(11):6079. https://doi.org/10.3390/ijms23116079
Kung T-H, Chao T-C, Xie Y-R, Pai M-C, Kuo Y-M, Lee GGC (2021) Neuroimage biomarker identification of the conversion of mild cognitive impairment to Alzheimer’s disease. Front Neurosci 15:584641. https://doi.org/10.3389/fnins.2021.584641
Lahat D, Adali T, Jutten C (2015) Multimodal data fusion: an overview of methods, challenges and prospects. Proc IEEE 103(9):1449–1477. https://doi.org/10.1109/JPROC.2015.2460697
Lao H, Zhang X (2022) Diagnose Alzheimer’s disease by combining 3D discrete wavelet transform and 3D moment invariants. IET Image Process. https://doi.org/10.1049/ipr2.12605
Lazli L, Boukadoum M, Ait Mohamed O (2019) Computer-aided diagnosis system of Alzheimer’s disease based on multimodal fusion: tissue quantification based on the hybrid fuzzy-genetic-possibilistic model and discriminative classification based on the SVDD model. Brain Sci 9(10):289. https://doi.org/10.3390/brainsci9100289
Lee J, Kim KW, Choi SH, Huh J, Park SH (2015) Systematic review and meta-analysis of studies evaluating diagnostic test accuracy: a practical review for clinical researchers-part II. statistical methods of meta-analysis. Korean J Radiol 16(6):1188–1196. https://doi.org/10.3348/kjr.2015.16.6.1188
Lee G, Kang B, Nho K, Sohn K-A, Kim D (2019) MildInt: deep learning-based multimodal longitudinal data integration framework. Front Genet 10:617. https://doi.org/10.3389/fgene.2019.00617
Lei B, Chen S, Ni D, Wang T (2016) Discriminative learning for Alzheimer’s disease diagnosis via canonical correlation analysis and multimodal fusion. Front Aging Neurosci 8:77. https://doi.org/10.3389/fnagi.2016.00077
Lei B et al (2020) Self-calibrated brain network estimation and joint non-convex multi-task learning for identification of early Alzheimer’s disease. Med Image Anal 61:101652. https://doi.org/10.1016/j.media.2020.101652
Li W, Lin X, Chen X (2020) Detecting Alzheimer’s disease Based on 4D fMRI: an exploration under deep learning framework. Neurocomputing 388:280–287. https://doi.org/10.1016/j.neucom.2020.01.053
Liu W, Zheng Z, Wang Z (2021a) Robust multi-focus image fusion using lazy random walks with multiscale focus measures. Signal Process 179:107850. https://doi.org/10.1016/j.sigpro.2020.107850
Liu DC et al (2021b) Detection of amyloid-beta by Fmoc-KLVFF self-assembled fluorescent nanoparticles for Alzheimer’s disease diagnosis. Chin Chem Lett 32(3):1066–1070
Liu M, Zhang J, Adeli E, Shen D (2018) Landmark-based deep multi-instance learning for brain disease diagnosis. Med Image Anal 43:157–168. https://doi.org/10.1016/j.media.2017.10.005
Liu S, Liu S, Cai W, Che H, Pujol S, Kikinis R, Feng D, Fulham MJ (2015) ADNI. Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s disease. IEEE Trans Biomed Eng 62(4):1132–1140. https://doi.org/10.1109/TBME.2014.2372011
Liu M, Cheng D, Wang K, Wang Y, Alzheimer’s Disease Neuroimaging Initiative (2018) Multi-modality cascaded convolutional neural networks for Alzheimer’s disease diagnosis. Neuroinform 16(3):295–308. https://doi.org/10.1007/s12021-018-9370-4
Lu D, Popuri K, Ding GW, Balachandar R, Beg MF (2018) Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s disease using structural MR and FDG-PET images. Sci Rep 8(1):5697. https://doi.org/10.1038/s41598-018-22871-z
Lu P, Hu L, Zhang N, Liang H, Tian T, Lu L (2022) A two-stage model for predicting mild cognitive impairment to Alzheimer’s disease conversion. Front Aging Neurosci 14:826622. https://doi.org/10.3389/fnagi.2022.826622
Lu D, Popuri K, Ding GW, Balachandar R, Beg MF (2018) Multiscale deep neural network based analysis of FDG-PET images for the early diagnosis of Alzheimer’s disease. Med Image Anal 46:26–34. https://doi.org/10.1016/j.media.2018.02.002
Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL (2007) Open access series of imaging studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cognitive Neurosci 19:1498–1507. https://doi.org/10.1162/jocn.2007.19.9.1498
Mathotaarachchi S et al (2017) Identifying incipient dementia individuals using machine learning and amyloid imaging. Neurobiol Aging 59:80–90. https://doi.org/10.1016/j.neurobiolaging.2017.06.027
Meng X et al (2022) Multi-modal neuroimaging neural network-based feature detection for diagnosis of Alzheimer’s disease. Front Aging Neurosci 14:3948–3964. https://doi.org/10.3389/fnagi.2022.911220
Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 151(4):264–269. https://doi.org/10.7326/0003-4819-151-4-200908180-00135
Muzammil SR, Maqsood S, Haider S, Damaševičius R (2020) CSID: a novel multimodal image fusion algorithm for enhanced clinical diagnosis. Diagnostics 10(11):904. https://doi.org/10.3390/diagnostics10110904
Nachappa DMN, Bojamma AM, Aparna MC (2018) A review on various fusion techniques in multimodal biometrics. Int J Eng Res Technol 4(21):1–8. https://doi.org/10.17577/IJERTCONV4IS21018
Ning Z, Xiao Q, Feng Q, Chen W, Zhang Y (2021) Relation-induced multi-modal shared representation learning for Alzheimer’s disease diagnosis. IEEE Trans Med Imaging 40(6):1632–1645. https://doi.org/10.1109/TMI.2021.3063150
Odusami M, Maskeliūnas R, Damaševičius R (2023) Pixel-level fusion approach with vision transformer for early detection of Alzheimer’s disease. Electronics. https://doi.org/10.3390/electronics12051218
Ortiz A, Lozano F, Gorriz JM, Ramirez J, Martinez Murcia FJ, Alzheimer’s Disease Neuroimaging Initiative (2018) Discriminative sparse features for Alzheimer’s disease diagnosis using multimodal image data. Current Alzheimer Res 15(1):67–79. https://doi.org/10.2174/1567205014666170922101135
Ou Y-N et al (2019) FDG-PET as an independent biomarker for Alzheimer’s biological diagnosis: a longitudinal study. Alzheimer’s Res Ther 11(1):57. https://doi.org/10.1186/s13195-019-0512-1
Pan J, and Wang S (2022) Cross-modal transformer GAN: a brain structure-function deep fusing framework for Alzheimer’s disease. arXiv, Accessed: Oct. 01, 2022. [Online]. Available: http://arxiv.org/abs/2206.13393
Pan J, Lei B, Wang S, Wang B, Liu Y, and Shen Y (2021a) DecGAN: decoupling generative adversarial network detecting abnormal neural circuits for Alzheimer’s disease. arXiv,. Accessed: Nov. 06, 2022. [Online]. Available: http://arxiv.org/abs/2110.05712
Pan J, Lei B, Shen Y, Liu Y, Feng Z, and Wang S (2021b) “Characterization Multimodal Connectivity of Brain Network by Hypergraph GAN for Alzheimer’s Disease Analysis,” in Pattern Recognition and Computer Vision, Cham, pp. 467–478. https://doi.org/10.1007/978-3-030-88010-1_39.
Peng J, Zhu X, Wang Y, An L, Shen D (2019) Structured sparsity regularized multiple kernel learning for Alzheimer’s disease diagnosis. Pattern Recogn 88:70–382. https://doi.org/10.1016/j.patcog.2018.11.027
Pham NTT et al (2022) Regional white matter hyperintensities in posterior cortical atrophy and logopenic progressive aphasia. Neurobiol Aging 119:46–55. https://doi.org/10.1016/j.neurobiolaging.2022.07.008
Qiu S, Chang GH, Panagia M, Gopal DM, Au R, Kolachalama VB (2018) Fusion of deep learning models of MRI scans, mini-mental state examination, and logical memory test enhances diagnosis of mild cognitive impairment. Alzheimer’s Dement: Diagn Assess Dis Monit 10:737–749. https://doi.org/10.1016/j.dadm.2018.08.013
Rajini KC and Roopa S (2017) A review on recent improved image fusion techniques,” in 2017 International conference on wireless communications, signal processing and networking (WiSPNET), pp. 149–153. https://doi.org/10.1109/WiSPNET.2017.8299737.
Rallabandi VPS, Tulpule K, Gattu M (2020) Automatic classification of cognitively normal, mild cognitive impairment and Alzheimer’s disease using structural MRI analysis. Inform Med Unlocked 18:100305. https://doi.org/10.1016/j.imu.2020.100305
Salvatore C, Cerasa A, Castiglioni I (2018) MRI characterizes the progressive course of AD and predicts conversion to Alzheimer’s dementia 24 months before probable diagnosis. Front Aging Neurosci 10:135. https://doi.org/10.3389/fnagi.2018.00135
S. Sarraf and G. Tofighi, “Classification of Alzheimer’s Disease using fMRI Data and Deep Learning Convolutional Neural Networks,” arXiv:1603.08631 [cs], Mar. 2016, Accessed: Apr. 06, 2021. [Online]. Available: http://arxiv.org/abs/1603.08631
Sayed I, Hassanien E, Nassef M, Pan S (2017) Alzheimer’s disease diagnosis based on moth flame optimization. In: Pan JS, Lin JCW, Wang CH, Jiang XH (eds) Genetic and evolutionary computing advances in intelligent systems and computing, vol 536. Springer, Cham, https://doi.org/10.1007/978-3-319-48490-7_35
Schouten TM et al (2016) Combining anatomical, diffusion, and resting state functional magnetic resonance imaging for individual classification of mild and moderate Alzheimer’s disease. NeuroImage: Clin 11:46–51. https://doi.org/10.1016/j.nicl.2016.01.002
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, and Batra D (2017) Grad-CAM: visual explanations from deep networks via gradient-based localization, In Proceedings of the IEEE international conference on computer vision, pp. 618–626.
Shahdoosti HR, Tabatabaei Z (2019) MRI and PET/SPECT image fusion at feature level using ant colony based segmentation. Biomed Signal Process Control 47:63–74. https://doi.org/10.1016/j.bspc.2018.08.017
Shankar K, Khanna A, Tanwar S, Rodrigues J et al (2019) Alzheimer detection using group grey wolf optimization-based features with convolutional classifier. Comput Electr Eng 77:230–243. https://doi.org/10.1016/j.compeleceng.2019.06.001
Shao W, Peng Y, Zu C, Wang M, Zhang D (2020) Hypergraph based multi-task feature selection for multimodal classification of Alzheimer’s disease. Comput Med Imaging Graph 80:101663. https://doi.org/10.1016/j.compmedimag.2019.101663
Sharma S, Mandal PK (2023) A comprehensive report on machine learning-based early detection of Alzheimer’s disease using multi-modal neuroimaging data. ACM Comput Surv 55(2):1–44. https://doi.org/10.1145/3492865
Shi J, Zheng X, Li Y, Zhang Q, Ying S (2018) Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer’s disease. IEEE J Biomed Health Inform 22(1):173–183. https://doi.org/10.1109/JBHI.2017.2655720
Singh D, Kaur M, Singh H (2018) Remote sensing image fusion using fuzzy logic and gyrator transform. Remote Sens Lett 9(10):942–951. https://doi.org/10.1080/2150704X.2018.1500044
Sintini I et al (2018) Regional multimodal relationships between tau, hypometabolism, atrophy, and fractional anisotropy in atypical Alzheimer’s disease. Hum Brain Mapp 40(5):1618–1631. https://doi.org/10.1002/hbm.24473
Song J, Zheng J, Li P, Lu X, Zhu G, Shen P (2021) An effective multimodal image fusion method using MRI and PET for Alzheimer’s disease diagnosis. Front Digital Health. https://doi.org/10.3389/fdgth.2021.637386
Suk H-I, Lee S-W, Shen D, The Alzheimer’s Disease Neuroimaging Initiative (2016) Deep sparse multi-task learning for feature selection in Alzheimer’s disease diagnosis. Brain Struct Funct 221(5):2569–2587. https://doi.org/10.1007/s00429-015-1059-y
Sun LM et al (2022) Blood-based Alzheimer’s disease diagnosis using fluorescent peptide nanoparticle arrays. Chin Chem Lett 33(4):1946–1950
Tong T, Gray K, Gao Q, Chen L, and Rueckert D. Nonlinear graph fusion for multi-modal classification of Alzheimer’s disease. In machine learning in medical imaging (pp. 77–84), 2016. Springer International Publishing. https://doi.org/10.1007/978-3-319-24888-2_10
Veitch DP et al (2022) Using the Alzheimer’s disease neuroimaging initiative to improve early detection, diagnosis, and treatment of Alzheimer’s disease. Alzheimer’s Dementia 18(4):824–857. https://doi.org/10.1002/alz.12422
Veshki FG, Ouzir N, Vorobyov SA and Ollila E, “Multimodal image fusion via coupled feature learning,” Signal Processing, vol. 200, p. 108637, Nov. 2022. https://doi.org/10.1016/j.sigpro.2022.108637.
Wang Y, Liu X, Yu C (2021) Assisted diagnosis of Alzheimer’s disease based on deep learning and multimodal feature fusion. Complexity 2021:e6626728. https://doi.org/10.1155/2021/6626728
Wang Z, Ma Y, Zhang Y (2023) Review of pixel-level remote sensing image fusion based on deep learning. Inform Fusion 90:36–58. https://doi.org/10.1016/j.inffus.2022.09.008
Xiao G, Bavirisetti DP, Liu G, and Zhang X (2020) Feature-Level Image Fusion,” in Image Fusion, Xiao G, Bavirisetti DP, Liu G, and Zhang X, Eds. Singapore: Springer, pp. 103–147. https://doi.org/10.1007/978-981-15-4867-3_3.
Xing X et al (2019) Dynamic spectral graph convolution networks with assistant task training for early mci diagnosis, in medical image computing and computer assisted intervention–MICCAI. Cham 2019:639–646. https://doi.org/10.1007/978-3-030-32251-9_70
Xu H, Zhong S, and Zhang Y, Multi-level fusion network for mild cognitive impairment identification using multi-modal neuroimages. Rochester, NY, Apr. 25, 2022. https://doi.org/10.2139/ssrn.4092904.
Yu S et al (2020) Multi-scale enhanced graph convolutional network for early mild cognitive impairment detection, in medical image computing and computer assisted intervention–MICCAI. Cham 2020:228–237. https://doi.org/10.1007/978-3-030-59728-3_23
Zhang T, Shi M (2020) Multi-modal neuroimaging feature fusion for diagnosis of Alzheimer’s disease. J Neurosci Methods 341:108795. https://doi.org/10.1016/j.jneumeth.2020.108795
Zhang Y, Wang S, Xia K, Jiang Y, Qian P (2021) Alzheimer’s disease multiclass diagnosis via multimodal neuroimaging embedding feature selection and fusion. Inform Fusion 66:170–183. https://doi.org/10.1016/j.inffus.2020.09.002
Zhang F, Li Z, Zhang B, Du H, Wang B, Zhang X (2019) Multi-modal deep learning model for auxiliary diagnosis of Alzheimer’s disease. Neurocomputing 361:185–195. https://doi.org/10.1016/j.neucom.2019.04.093
Zheng X, Shi J, Li Y, Liu X, and Zhang Q (2016) Multi-modality stacked deep polynomial network based feature learning for Alzheimer’s disease diagnosis, in 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 851–854. https://doi.org/10.1109/ISBI.2016.7493399.
Zuo Q, Lei B, Shen Y, Liu Y, Feng Z, Wang S (2021) Multimodal representations learning and adversarial hypergraph fusion for early Alzheimer’s disease prediction”, in pattern recognition and computer vision. Cham. https://doi.org/10.1007/978-3-030-88010-1_40
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Odusami, M., Maskeliūnas, R., Damaševičius, R. et al. Machine learning with multimodal neuroimaging data to classify stages of Alzheimer’s disease: a systematic review and meta-analysis. Cogn Neurodyn 18, 775–794 (2024). https://doi.org/10.1007/s11571-023-09993-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11571-023-09993-5