Abstract
In this study, we investigated two distinct new phylogenetic lineages of root-colonizing dark septate endophytic fungi colonizing wheat (Triticum aestivum) roots from a long-term agricultural experimental site in Hungary. According to four-locus (internal transcribed spacer, partial large and small subunit regions of nuclear ribosomal DNA, and partial translation elongation factor 1-alpha) phylogenetic analyses, the isolates belong to the Lentitheciaceae and Didymosphaeriaceae of the Pleosporales (Dothideomycetes). We studied the morphology and culture characteristics of the strains. We carried out in vitro resynthesis pot experiments with their original hosts and found no overall negative effect of the inoculation with different isolates of the new taxa. One of the lineages belonged to the genus Poaceascoma (Lentitheciaceae) and represented a novel species described here as Poaceascoma zborayi. We could describe conidia-like structures from this species. Isolates of the other lineage represented a monotypic novel genus in the Didymosphaeriaceae. Accordingly, the new genus, Agrorhizomyces, represented by the species A. patris, is introduced. Sterile, globose structures resembling immature sporocarps were detected. Sequence similarity searches indicated that P. zborayi might be widely distributed, while no sequence similar to A. patris was found outside the sampling area.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants live in association with various microorganisms, including fungal endophytes, both in natural and agricultural environments. Fungal endophytes colonize above- and below-ground plant tissues without causing disease symptoms to their hosts, at least during some period of their life cycle (Petrini 1991; Saikkonen et al. 1998; Vandenkoornhuyse et al. 2002; Mandyam and Jumpponen 2005; Schultz and Boyle 2005; Rodriguez et al. 2009). A form-group of root colonizing fungi accommodates dark septate endophytes (DSEs, Jumpponen and Trappe 1998), classified also as class 4 endophytes (Rodriguez et al. 2009), that form melanized septate hyphae. DSEs are common in root-associated fungal communities in several biomes and climatic regions, yet several lineages represent undescribed taxa, and the ecological functions of DSEs remain elusive (Mandyam and Jumpponen 2005; Porras-Alfaro and Bayman 2011; Sieber and Grünig 2013; Knapp et al. 2018, 2019).
The orders Helotiales (Leotiomycetes) and Pleosporales (Dothideomycetes) are the two orders accommodating most DSEs (Andrade-Linares and Franken 2013; Knapp et al. 2022, 2015; Grünig et al. 2011; Newsham 2011; Sieber and Grünig 2013; Jumpponen et al. 2017). However, other DSEs can also be found in the Hypocreales (Sordariomycetes) and Chaetothyriales (Eurotiomycetes) (Sieber and Grünig 2013; Jumpponen et al. 2017; Maciá-Vicente et al. 2018). Although narrow host specificity in DSE-plant interactions is not yet proven, different DSE communities are seen in trees and forest ecosystems and grasses and grasslands. Species belonging to the order Pleosporales are prevalent DSEs of the latter habitats, comprising a plethora of grass-root endophytes (Zhang et al. 2012; Jumpponen et al. 2017) that are present in natural (Rudgers et al. 2022) and agricultural ecosystems (Gdanetz and Trail 2017). The number of formally described DSE species is continuously increasing, and the vast majority of DSEs were isolated from natural ecosystems. For example, during the last decade, 14 new DSE species have been described from natural semiarid grassland ecosystems in Hungary (Knapp et al. 2015, 2022; Ashrafi et al. 2018; Crous et al. 2019, 2021). On the other hand, we should bear in mind that non-pathogenic fungi and endophytes in agricultural environments are understudied.
As more than half of the area of Hungary represents agronomic areas and more than 80% of those areas are croplands (Hungarian Central Statistical Office), it is important to gain more information about the non-pathogenic fungi of farmland habitats. We have studied a unique, long-term agricultural experimental area near Martonvásár, Hungary that is based on wheat (Triticum aestivum) and maize (Zea mays) monocultures and was similarly managed for more than 60 years (for detailed site description, see Mayer et al. 2019; Ujvári et al. 2020; Megyes et al. 2021). During the characterization of fungal root endophytes of different parcels in these experiments, we gained hundreds of different isolates. The initial DNA barcoding using ITS (internal transcribed spacer) of nrDNA sequences revealed that several isolates from wheat represent two novel pleosporalean lineages distinct from the known species in the genus Poaceascoma (Lentitheciaceae) and the known genera in the Didymosphaeriaceae. In this study, we aimed at (i) determining the phylogenetic position of these lineages and clarify the taxonomy by providing formal descriptions of novel taxa and (ii) conducting in vitro resynthesis experiments to gain insights into their interaction with host plants.
Materials and methods
Sampling and isolation of fungal strains
Root samples of wheat were collected in experimental fields located near Martonvásár, Hungary (N47˚ 16′ 36.23′′; E18˚ 47′ 39.65′′). Root samples were collected on 08.07.2019. Healthy wheat roots were surface sterilized and processed as described in Knapp et al. (2012). We collected 32 isolates representing two distinct lineages (11 and 21 isolates). A selection of ten isolates were chosen for further analyses (Table 1), based on macroscopical characteristics on different culture media and sporulation inducing experiments. Four isolates representing a lineage in the genus Poaceascoma (Lentitheciaceae) and six Didymosphaeriaceae were chosen for molecular phylogenetic analysis (Tables 2 and 3). Three isolates of the former six of the latter were chosen for the resynthesis experiments and further sporulation generation experiments on oatmeal agar. Holotypes consisted of lyophilized cultures and were deposited as metabolically inactive samples in the herbarium of the Hungarian Natural History Museum, Budapest (BP) under the accession numbers BP112746 and BP112747. Ex-type and other cultures investigated in this study were deposited in the culture collection of the Westerdijk Fungal Biodiversity Institute (CBS 151097 and CBS 151043); nomenclatural novelties and descriptions were deposited in MycoBank (www.MycoBank.org, Crous et al. 2004).
DNA extraction and amplification
Genomic DNA was extracted from fungal mycelia using the NucleoSpin Plant II kit (using PL1, Macherey–Nagel, Germany) following the manufacturer’s instructions. Four loci were amplified and sequenced: internal transcribed spacer (ITS), partial 18S small subunit (nc SSU rDNA), and partial 28S large subunit (nc LSU rDNA) of the nrDNA, and partial translation elongation factor 1-alpha gene (TEF1–α). The following primers were used for amplification and sequencing: for ITS, ITS1F/ITS4 (White et al. 1990; Gardes and Bruns 1993); for nc SSU rDNA, NS1/NS4 (White et al. 1990); for nc LSU rDNA, LR0R/LR5 (Rehner and Samuels 1994; Vilgalys and Hester 1990); for TEF1–α, EF1-983/EF1-2218R (Rehner and Buckley 2005). The sequences were compiled from electropherograms using the Pregap4 and Gap4 software packages (Staden et al. 2000) and Sequencher 5.4 (GeneCodes Corporation, Ann Arbor, Michigan, USA), and deposited in GenBank (PP264924–PP265020 and PP273237–PP273267; Tables 1, 2 and 3). The obtained sequences were compared with the accessions in the National Center for Biotechnology Information database (NCBI, http://www.ncbi.nlm.nih.gov/Blast.cgi) using the BLASTn search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al. 1990). We also compared the ITS sequences with two of our unpublished datasets: one is a pilot fungal NGS metabarcoding dataset from the maize monocultures of the sampling site (see details on bacterial communities in Megyes et al. 2021), and the other is the fungal metabarcoding data from a long-term ecological experiment form the semiarid sandy grasslands of Fülöpháza, Hungary (Vajna et al. 2021).
Phylogenetic analyses
We combined and aligned the sequences of different loci with those from representative taxa in GenBank using the online version of MAFFT 7 (Katoh and Standley 2013) and the E-INS-i method. The alignments were examined and edited using MEGA 7 (Kumar et al. 2016). Two multi-locus family-level datasets for Lentitheciaceae (90 taxa) (Table 2) and Didymosphaeriaceae (110 taxa) (Table 3) were used for molecular phylogenetic analyses. Sequences highly similar to data acquired from the comparison with the accessions NCBI database using the BLASTn search (Altschul et al. 1990) and strains of representative species of the families Lentitheciaceae according to Calabon et al. (2021), Liu et al. (2022), Rajeshkumar et al. (2023), Hyde et al. (2021), and Didymosphaeriaceae according to Samarakoon et al. (2020b), Yuan et al. (2020), Crous et al. (2022), Verkley et al. (2014) and Valenzuela-Lopez et al. (2018) were involved into the phylogenetic analyses. For both phylogenetic analyses, Corynespora cassiicola (CBS 100822) and Corynespora smithii (CABI5649b) served as outgroups. Bayesian inference (BI) analyses were performed with MRBAYES 3.1.2 (Ronquist and Huelsenbeck 2003) with a GTR + G substitution model for all nucleotide partitions. Four Markov chains were run for 10,000,000 generations, sampling every 1,000 generations with a burn-in value set at 4,000 sampled trees. Maximum likelihood (ML) phylogenetic analysis was carried out with RAXMLGUI 1.3 (Silvestro and Michalak 2012; Stamatakis 2014). The GTR + G nucleotide substitution model was also used for nucleotide partitions with ML estimation of base frequencies. ML bootstrap (BS) analysis with 1,000 replicates was used to test the support of the branches. Another dataset was created with only ITS sequences for the genus Poaceascoma, including five sequences most similar to Poaceascoma zborayi based on a BLASTn search in NCBI database. The sequences were aligned and analysed as described above using Setoseptoria arundelensis (MFLUCC 17–0759) and Setoseptoria englandensis (MFLUCC 17–0778) as outgroups. Phylogenetic trees were visualized and edited in MEGA 7 (Kumar et al. 2016) and deposited in Figshare (doi: 10.6084/m9.figshare.25164665).
Fungal morphology and sporulation
Potato dextrose and malt extract agar (PDA, MEA, VWR International, Belgium) plates were inoculated with colony plugs (5 mm diameter) in Petri dishes (5 cm diameter). Growth rate and colony characteristics were recorded in cultures grown for three weeks at 22 °C. To induce sporulation, isolates were cultured on autoclaved pine needles and stinging nettle stems laid on water agar (WA) in Petri dishes (9 cm diameter). The isolates were also cultured on WA media supplemented with minced vegetables (carrot, turnip, celery, and kohlrabi) at a pH of 3.5 (pH meter: Consort C830). Cultures were also grown on oatmeal agar (OA) in Petri dishes (5 cm diameter). The oatmeal agar contained 500 ml water, 15 g thin-rolled oats and 7.5 g agar. The ingredients were mixed and autoclaved following Gooding and Lucas (1959). The isolates were growing on these media at 22 °C for three months. Fungal structures and pieces of colonies were positioned and frozen onto a Peltier-cell cooled stage in distilled water mixed with glycerol. Sections of 30–50 μm thickness were cut with a Reichert microtome (Reichert, Austria) with steel microtome knife, and the sections were collected and mounted in distilled water mixed with glycerol. Morphological characteristics of the fungal structures were examined using a light microscope with differential interference contrast (DIC) optics and a Nikon Eclipse 80i microscope equipped with a Spot 7.4 Slider camera (Diagnostic Instruments, Inc.). Measurements and photographs were made using structures mounted in either water or in polyvinyl-lactoglycerol (PVLG).
Resynthesis experiment
An artificial in vitro resynthesis pot experiment was conducted for testing Poaceascoma isolates: BDT06, BDT15 and BDT33 and Didymosphaeriaceae isolates: BAT15, BAT20, BAT41, BBT01, BBT16, BBT24. In the experiment, 1.5 dl pots were filled with a twice autoclaved mixture of original soil from Martonvásár, sand, and zeolite (1:1:1). Grains of wheat were surface sterilized, allowed to germinate and each seedling was inoculated with five 5-mm fungal plugs, that were placed into the soil near the roots of seedlings. The plants were grown for eight weeks at 22 °C under a 14 h light:10 h dark cycle. Five replicates were used for each fungal isolate. The control pots were “inoculated” with five 5-mm plugs of the sterile medium. Altogether 50 plants were assessed. At the end of the inoculation experiments, the plants were harvested, and the substratum was carefully removed from the surface of the root system. Shoots were separated from the roots, and both were dried at 50 °C till constant weight, and then the shoot and root dry biomass was measured. Before drying, an equal amount of root segments was collected from each plant for microscopy analysis. To visualize the hyphae within the roots we used two methods. A wheat germ agglutinin (WGA) conjugate, WGA-Alexafluor®488 (Thermo Fisher Scientific, Lithuania) was used that stains chitin and is widely used as a fluorescent dye for fungal cell walls (Németh et al. 2022). The other method applied aniline blue as a stain for in planta visualization of fungal endophytes (Andrade-Linares et al. 2011; Knapp et al. 2019). The slides were examined on the Nikon microscope and setup described above and with excitation and emission filters for visualization of Alexa Fluor 488 probe. To test the effect of inoculation, one-way analysis of variance (ANOVA) was applied with a Tukey’s test for post-hoc analysis to identify the differences in dry biomass among plants inoculated by different isolates. For statistical analyses the GraphPad Prism 9.5.0 software was used.
Results
Molecular phylogeny
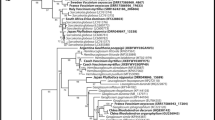
The four-loci phylogenetic analysis of Lentitheciaceae placed strains BDT06, BDT15, BDT32 and BDT33 into a clade with six previously described Poaceascoma species with high support (ML-BS = 93, B-PP = 0.99) (Fig. 4). Based on the phylogenetic analysis, the position of Poaceascoma within Lentitheciaceae is ambiguous, however, it grouped together with Stagonospora and Setoseptoria species with a moderate support. Poaceascoma taiwanense represents a distinct branch, and strains of the type species of Poaceascoma, P. helicoides, clustered with an ambiguously characterized strain P. lochii BRIP 71546 (ML-BS = 100, B-PP = 1.00). Three species, P. filiforme, P. halophilum, and our isolates grouped together (ML-BS = 71, B-PP = 0.99), and the ex-type strain, P. halophilum (MFLUCC 15–0949), formed a well-supported clade with those from wheat roots (ML-BS = 81, B-PP = 0.99). The four isolates form a strongly supported clade (ML-BS = 95, B-PP = 0.99). Relatedness of isolates BAT15, BAT20, BAT41, BBT01, BBT16 and BBT24 with Didymosphaeriaceae is highly supported (ML-BS = 100, B-PP = 1) (Fig. 5). Three clades, consisting of genera Spegazzinia and Dictyoarthrinium, and a distinct lineage represented by our isolates from Hungary, formed a strongly supported (ML-BS = 86, B-PP = 1) basal group in Didymosphaeriaceae. Each of these three clades has full support (ML-BS = 100, B-PP = 1), including the one comprising six isolates with identical sequences from wheat.
The 32 isolates, of which ten isolates were studied in detail in this study, formed two distinct lineages – these clades are considered as novel species in the genus Poaceascoma (Lentitheciaceae) and a novel species belonging to a new monotypic genus in Didymosphaeriaceae, and those taxa are formally described here.
Taxonomy
Poaceascoma zborayi Imrefi, D.G. Knapp & Kovács, sp. nov. — MycoBank MB852022; Figs. 1 and 4.
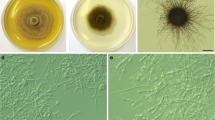
Colony and microscopic images of Poaceascoma zborayi strain BDT15 (CBS 151097). a On potato dextrose agar (PDA) incubated at 22 °C in darkness b Globose structure formed on oatmeal agar (OA) after three weeks at room temperature c Conidia-like structures formed on oatmeal agar (OA) after three weeks at room temperature. d Conidia-like structures formed on oatmeal agar (OA) after three weeks at room temperature. Scale bars: b–c 200 μm; d100 μm
Etymology: We name the species in honour of Géza Zboray (1941–2023), an outstanding teacher at the Institute of Biology, Eötvös Loránd University (Budapest, Hungary), who had considerable influence on generations of Hungarian biologists all around the country, including all here involved authors.
Typification: Hungary: Martonvásár, isolated from healthy roots of wheat collected from agricultural fields, N47˚ 16′ 36.23′′; E18˚ 47′ 39.65′′, July 2019, I. Imrefi, a dried biologically inert agar culture (holotype BP112746, deposited under the barcode HNHM-MYC 033463; ex-type culture BDT15 = CBS 151097). GenBank: ITS = PP264928; nc LSU rDNA = PP264982; nc SSU rDNA = PP265014; TEF1–α = PP273262.
Diagnosis: Based on the phylogenetic analyses Poaceascoma zborayi differs from the taxa phylogenetically analysed in Fig. 4 by unique fixed alleles in the TEF1-α and nc ITS rDNA loci, identified based on alignments of separate loci deposited at Figshare repository (doi: 10.6084/m9.figshare.25164665): TEF1-α positions: 629 (T), 695 (T), 941 (T); nc ITS rDNA positions: a unique nucleotide motif at 350–363 (CGTGCGTTGGACCT). Additionally to those, Poaceascoma zborayi differs from congeneric Poaceascoma species by unique fixed alleles in the TEF1-α, nc ITS rDNA and nc LSU rDNA identified based on alignments of separate loci deposited at Figshare repository (doi: 10.6084/m9.figshare.25164665): TEF1-α positions: 97 (C), 244 (T), 374 (T), 389 (T), 407 (A), 416 (T), 443 (T), 479 (G), 482 (C), 656 (T), 731 (T), 758 (C), 779 (C), 791 (C), 803 (T), 824 (T), 833 (T), 854 (C); nc ITS rDNA positions: 36 (A), 81 (C), 82 (DEL), 87 (T), 103 (G), 120 (A), a unique nucleotide motif at 331–369 (CTGGGTGTTGTCCCGCCTCGTGCGTTGGACCTCGCCCG), 439 (C), 468 (T), 489 (A), 495 (A), 511 (C); nc LSU rDNA positions: 158 (T), 375 (T).
Additional specimens examined: Hungary: Martonvásár, roots of wheat collected from agricultural fields N47˚ 16′ 36.23′′; E18˚ 47′ 39.65′′, July 2019, I. Imrefi (BDT06); ibid. (BDT32); ibid. (BDT33); ibid. (BDT05); ibid. (BDT07); ibid. (BDT14); ibid. (BDT46); ibid. (BDT47); ibid. (BDT49); ibid. (BDT50). (Table 1).
Description: Poaceascoma zborayi is a dark septate endophyte colonizing wheat roots, also in in vitro resynthesis experiments, with no negative effects on the host. The isolates of the species showed almost the same morphology and dark colour on all the media tested. Colonies moderately slow-growing at 22 °C, on PDA covered the 5 cm Petri dishes, on MEA reaching 41 mm diam in 21 d. Colonies on PDA brownish grey with dark grey marginal zone and flat, with sparse to dense aerial mycelium, radially striate as the centre is lighter brown than the younger part of the colony, with dark exudates in the media. On MEA, colonies dark brown with a pale grey marginal zone and flat, with sparse aerial mycelium and radially striate, without secreted exudate droplets (Fig. 1a). Conidia-like structures produced by BDT15 on OA medium (Fig. 1c, d): conidiophore-like structures formed by branched, segmented hyaline hyphae, often found in groups. Conidia-like structures emerging on these structures were observed, brownish-red, rough-walled, warted, having variable sizes (20–40 μm). Immature, globose structures also formed by BDT15 (CBS 151097) on OA (500–800 µm diam) (Fig. 1b). Isolates BDT15 and BDT33 also formed hyphal loops on OA. The ITS blast search (08/01/2024) found several GenBank entries with more than 99% similarity with the ITS of P. zborayi, for example: MK808094 (Rudgers et al. 2022), OM106729 (Beschoren da Costa et al. 2022), KJ188723 (Luo et al. 2014), DQ420978 (Waldrop et al. 2006), MT683270 (Fors et al. 2020). Based on the ITS search in two of our unpublished fungal metabarcoding datasets, a fungus with 99.5% similarity to P. zborayi was found in the soil samples of maize monoculture on Martonvásár, but P. zborayi was not found in the soil from natural sandy grassland near Fülöpháza.
Agrorhizomyces Imrefi, D.G. Knapp & Kovács, gen. nov. — MycoBank MB852052; Figs. 2 and 5.
Colonies with different morphology and microscopic images of Agrorhizomyces patris a–c Colonies incubated on potato dextrose agar at 22 °C in darkness. a Isolate BBT01 (CBS 151043) b Isolate BAT20 c: Isolate BAT15 d Sterile, globose structure formed by isolate BBT01 after three weeks at room temperature on oatmeal agar. Scale bar: 200 μm
Etymology: Agro (from the Greek word agros, meaning field, a reference to the sampling site of the isolates, also to the Agricultural Institute of Centre for Agricultural Research) + rhizo (from the Greek word rhiza, meaning root, referring to the fact that the isolates were collected from surface sterilized roots).
Type species: Agrorhizomyces patris Imrefi, D.G. Knapp & Kovács.
Notes: The genus Agrorhizomyces contains one root endophytic species. Isolates can be collected from surface sterilized roots and can be cultured and maintained on general media.
Phylogenetically, the monotypic Agrorhizomyces groups within the Didymosphaeriaceae with full support.
Agrorhizomyces patris Imrefi, D.G. Knapp & Kovács, sp. nov. — MycoBank MB852054; Figs. 2 and 5.
Etymology: The species was named “patris” in honor and memory of Ildikó Imrefi’s father.
Typification: Hungary: Martonvásár, isolated from healthy roots of wheat collected from agricultural fields N47˚ 16′ 36.23′′; E18˚ 47′ 39.65′′, July 2019, I. Imrefi, a dried biologically inert agar culture (holotype BP112747, deposited under the barcode HNHM-MYC 033464; ex-type culture BBT01 = CBS 151043). GenBank: ITS = PP264945; nc LSU rDNA = PP264967; nc SSU rDNA = PP264999; TEF1–α = PP273247.
Diagnosis:
Based on the phylogenetic analyses Agrorhizomyces patris differs from the taxa phylogenetically analysed in Fig. 5 by unique fixed alleles in the TEF1-α, nc ITS rDNA and nc LSU rDNA loci identified based on the alignments of separate loci deposited at Figshare repository (doi: 10.6084/m9.figshare.25164665): TEF1-α position: 218 (T); nc ITS rDNA positions: a unique nucleotid motif at 226–232 (GTATACC), 246 (C), 482–484 (TAA), a unique nucleotid motif at 561–595 (AGAGTAGGCGGTTGCTCGAGGCTTT); nc LSU rDNA positions: 290 (C), 297 (G), 369 (G), 375–376 (AA), 426–427 (TC), 564 (C).
Additional specimens examined: Hungary: Martonvásár, roots of wheat collected from agricultural fields N47˚ 16′ 36.23′′; E18˚ 47′ 39.65′′, July 2019, I. Imrefi (BAT15); ibid. (BAT20); ibid. (BAT41); ibid. (BBT16); ibid. (BBT24); ibid. (BAT14); ibid. (BAT17); ibid. (BAT19); ibid. (BAT21); ibid. (BAT25); ibid. (BAT39); ibid. (BAT40); ibid. (BBT01); ibid. (BBT02); ibid. (BBT04); ibid. (BBT13); ibid. (BBT15); ibid. (BBT24); ibid. (BBT41); ibid. (BBT42); ibid. (BBT43); ibid. (BBT55).
Description: Agrorhizomyces patris is a dark septate endophyte colonizing wheat roots, also in in vitro resynthesis experiments, with no or slightly negative effects to the host. The isolates show variant colony morphologies and colours and growing characteristics on different media (Fig. 2a–c).Colonies moderately fast-growing on PDA, growing approximately 0.7 cm a week, on other examined culture media (MEA) at 22 °C covered 5 cm Petri dishes in 21 d. Colonies on PDA pale brown grey with creamy marginal zone, colony surface slightly elevated at the centre, with dense aerial mycelium, radially striate, with sparse secreted exudate droplets on the surface of the mycelia. On MEA, colonies dark brown-grey with a greyish inner zone, colony surface slightly elevated at the centre, with dense aerial mycelium, radially striate, with sparse secreted exudate droplets on the surface of the mycelia (Fig. 2a–c). Sexual morph unknown. Globose structures formed by BBT01 (CBS 151043) on OA medium (Fig. 2d), consisting of wall, approximately 200 μm thick surrounding a cavity. These structures are strongly pigmented, resembling immature sporocarps with variable sizes (1.5–2 mm diam). Sporulation was not observed. The ITS BLASTn search (08/01/2024) in GenBank resulted only three unpublished environmental sequences with ~ 93% similarity with the ITS sequence of the A. patris isolate BBT01. These three almost identical sequences were: MF569188, KX192630, ON696029. Based on the ITS search in two of our unpublished fungal metabarcoding datasets a fungus with 100% similarity to A. patris was found in the soils of maize monoculture at Martonvásár, but A. patris was not found in the soil from a natural grassland near Fülöpháza.
Resynthesis experiment
In the resynthesis experiment, all nine isolates were able to colonize the wheat roots, forming characteristic extra- and intraradical hyphal structures and microsclerotia (Online Resource 1 b, c). Intraradical hyphae of both species were septate (Online Resource 1 a, b, d). Intraradical hyphae of Agrorhizomyces patris were predominantly hyaline and could be stained with the WGA-Alexafluor®488 fluorescent dye (Online Resource 1c). Those of Poaceascoma zboray were melanized and could not be visualized by WGA-Alexafluor®488 fluorescent dye (Online Resource 1a, b). The presence of the isolates effected the biomass of wheat in the pot culture experiment based on one-way ANOVA (p < 0.01). However, based on the results of Tukey’s test, only one of the isolates effected the biomass significantly (p < 0.05) compared to the control plant: BBT16 caused a decrease of shoot and root dry weight (Fig. 3).
Discussion
Here we introduced and formally described two novel pleosporalean DSE species, Poaceascoma zborayi and Agrorhizomyces patris isolated from healthy wheat roots from an agricultural field. There are more than 40 well-identified and investigated pleosporalean species considered as DSEs in genera Acrocalymma, Alternaria, Alfoldia, Aquilomyces, Curvularia, Darksidea, Delitchia, Drechslera, Flavomyces, Fuscosphaeria, Kiskunsagia, Laburnicola, Murispora, Periconia, Polydomus, Posidoniomyces, Setophoma and numerous further undescribed lineages belonging to families mainly of suborders Pleosporineae and Massarineae (Knapp et al. 2012, 2015, 2018, 2022; Zhang et al. 2012; Andrade-Linares and Franken 2013; Sieber and Grünig 2013; Knapp and Kovács 2016; Jumpponen et al. 2017; Vohník et al. 2019; Crous et al. 2019; Pereira et al. 2019; Crous et al. 2021; Pintye and Knapp 2021; Romero-Jiménez et al. 2022; Ashrafi et al. 2023). The majority of the known pleosporalean DSE species originated from natural ecosystems. However, one of the best-known and characterized DSE species, Periconia macrospinosa (Periconiaceae, Pleosporales) (Mandyam et al. 2010; Knapp et al. 2018) was originally isolated and described from roots of Sorghum vulgare (Lefebvre et al. 1949). Later it was regularly isolated from winter wheat roots (Hall 1986). The DSE species Murispora kazachstanica (Crous et al. 2021) and Laburnicola radiciphila (Knapp et al. 2022), were isolated from gramineous crops.
The pleosporalean family Lentitheciaceae was introduced by Zhang et al. (2009) based on multi-gene phylogeny. Lentitheciaceae currently accommodates 18 genera (see Calabon et al. 2021; Liu et al. 2022; Yang et al. 2022). Most of these genera comprise species characterized by lenticular to globose ascomata. Two genera (Phragmocamarosporium and Towyspora) accommodate only anamorphic species. Both are characterized by coelomycetous asexual morphs (Liu et al. 2022) (Figs. 4 and 5).
a Maximum Likelihood tree of concatenated ITS, nc LSU rDNA, nc SSU rDNA and TEF1-α sequences of representative species and genera of the Lentitheciaceae tree including Poaceascoma species. ML bootstrap support values (≥ 70) are shown before slashes or above branches, Bayesian posterior probabilities (≥ 0.90) are shown after slashes or below branches. Highlighted margin sections indicate names of taxa, the here described Poaceascoma zborayi is in bold. Corynespora cassiicola (CBS 100822) and Corynespora smithii (CABI5649b) served as outgroups. The scale bar indicates expected changes per site per branch. b Maximum Likelihood (RAxML) tree of ITS sequences of Poaceascoma species and P. zborayi isolates with bootstrap values and scale bar as described above. Setoseptoria arundelensis (MFLUCC 17–0759) and Setoseptoria englandensis (MFLUCC 17–0778) served as outgroups
Maximum Likelihood tree of concatenated ITS, nc LSU rDNA, nc SSU rDNA and TEF1-α sequences of representative species and genera of the Didymosphaeriaceae tree including Agrorhizomyces patris. ML bootstrap support values (≥ 70) are shown before slashes or above branches, Bayesian posterior probabilities (≥ 0.90) are shown after slashes or below branches. Highlighted margin sections indicate the names of taxa, the here described A. patris is in bold. Corynespora cassiicola (CBS 100822) and Corynespora smithii (CABI5649b) served as outgroups. The scale bar indicates expected changes per site per branch
The initially monoptypic genus Poaceascoma was introduced with the species P. helicoides occurring on dead stems and roots of Digitaria sanguinalis (Poaceae) collected in a natural terrestrial habitat (Phookamsak et al. 2015). Later, five additional species were described. All six species were described based on their sexual morphs. Typically, ascomata are known from natural substrates (Poaceascoma helicoides, P. aquaticum, P. halophila, and P. taiwanense), while those of P. filiforme were described from oatmeal agar (Crous et al. 2020). These ascomata differed in several characteristics but were semi-immersed to erumpent with filiform, generally septate and spirally twisted ascospores. In phylogenetic analyses of different combinations of DNA loci, generally ITS, nc LSU rDNA, nc SSU rDNA, TEF1-α and RPB2, Poaceascoma species grouped together in well supported clades (Phookamsak et al. 2015; Luo et al. 2016; Hyde et al. 2017, 2018; Crous et al. 2020).
Further five Poaceascoma species (P. aquaticum, P. halophilum, P. taiwanense, P. filiforme, P. lochii) were described and reported as saprobic fungi from different gramineous plant materials (Luo et al. 2016; Hyde et al. 2017, 2018; Crous et al. 2020; Boonmee et al. 2021; Tan et al. 2021).
Here we introduced the seventh species in the genus Poaceascoma, P. zborayi, also collected from a gramineous plant species (Triticum aestivum) supporting the association of the genus to Poaceae plants. Asexual morph of none of the five already described Poaceascoma species was determined. Here we could observe the production of two different structures: P. zborayi (isolate BTD15) formed globose immature sporocarps that we could neither identify as ascomata nor pycnidia (Fig. 1b). They were larger (500–800 µm diam) than most of the ascomata of known Poaceascoma species (Hyde et al. 2018). Brownish-red, rough-walled, warted, segmented conidium-like structures were repeatedly observed, emerging from conidiophore structures with variable sizes (20–40 μm) (Fig. 1c, d). Such asexual propagules were neither described for other Poaceascoma species nor in other members of the family before.
Although the majority of Poaceascoma species and materials were collected from dead plant shoot tissues, Phookamsak et al. (2015) noted that ascomata can also be found on roots of Digitaria sanguinalis and observed P. helicoides as saprobic on grass culms and roots. Since P. zborayi is characterized as a root endophyte, not causing symptoms and with no negative effect in in vitro experiments, we might hypothesize that Poaceascoma species could have an endophytic lifestyle beside being collected as saprobes on plant debris. The ITS blast search (08/01/2024) showed several GenBank entries with more than 99% similarity with the ITS of P. zborayi illustrating its wide distribution: Rudgers et al. (2022) collected two isolates (e.g. MK808094) from healthy roots of the foundation grass Schizachyrium scoparium in North American plains. Numerous sequences (e.g., OM106729) were originated from the USA from roots of switchgrass (Panicum virgatum) in long-term bioenergy research sites in Michigan and Wisconsin (Beschoren da Costa et al. 2022). Luo et al. (2014) collected an isolate (KJ188723) from the roots of a dominant grass species in temperate pine barrens in New Jersey, USA. Waldrop et al. (2006) gained 100% similar uncultured clone sequences (e.g. DQ420978) from grass-dominated experimental plots in Minnesota, USA. The blast of ITS2 resulted additional 100% similar hits: sequence (ON696510) originating from the soil of an experimental site at Richmond, NSW, Australia (Hassan et al. 2022) and sequence (MF568887) originating from soil samples Bloomington-Normal, IL USA (Beck 2017). Based on similar sequences, other Poaceascoma isolates were found in various geographic areas also associated with agricultural plants. For instance, Raza et al. (2019) collected P. helicoides isolates from the gramineous crop sugarcane (Saccharum officinarum) in China. Several Poaceascoma isolates (e.g., A122, MT683270) were collected also from healthy roots of sugarcane in Brazil (Fors et al. 2020). These isolations of different Poaceascoma species from healthy roots of agricultural plants further strengthen the hypothesis that members of this genus could have an endophytic lifestyle.
The non-pathogenic association of the genus to the Poaceae was hypothesized by Wu et al. (2022). During their experiments on a chemical fertilizer reduction system using substitution by organic material inputs, a member of the genus Poaceascoma was a key player in significant changes of the potential fungal functions in co‑occurrence network patterns of bacterium‑fungus‑nematode communities. Poaceascoma zborayi colonized the host plant in our resynthesis experiments, causing no symptoms and without negative effect on the biomass of shoot and root, either. Therefore, we consider this species as a root endophyte and not a pathogen. Still, information on the functional role of Poaceascoma species remains limited.
The Didymosphaeriaceae represents a well-supported clade within the Massarineae (Tanaka et al. 2015; Yuan et al. 2020). Ariyawansa et al. (2014) synonymized Montagnulaceae under Didymosphaeriaceae, accommodating 39 genera including our new genus Agrorhizomycota (Wijayawardene et al. 2014, 2022; Liu et al. 2022 Tanaka et al. 2015; Wanasinghe et al. 2016; Yuan et al. 2020, Ren et al. 2022). More than two third of the genera in the family were introduced based on their sexual morphs. Their ascomata are generally globose to sub-globose, have a central ostiole, and peridia with several layers of lightly pigmented to dark brown or black cells of angular texture. The asci are 2–4 or 8-spored, comprising 1–2-seriate, overlapping, ellipsoid or oblong, 1–3-septate or muriform ascospores (Ren et al. 2022). Other genera were described by asexual morphs, such as Alloconiothyrium, Dictyoarthrinium, Paraconiothyrium, and Spegazzinia were introduced based on only their asexual morphs characters (Ren et al. 2022).
In the present study, the multi-locus analyses showed that Agrorhizomyces grouped with Spegazzinia and Dictyoarthrinium, forming a basal clade in Didymosphaeriaceae. Spegazzinia was established in 1880 by Saccardo based on S. ornate. Micromorphology supports its classification, in the Apiosporaceae (Sordariomycetes) (Samarakoon et al. 2020a). Based on a multi-locus phylogeny using sequences of S. deightonii and S. tessarthra, Tanaka et al. (2015) placed Spegazzinia in Didymosphaeriaceae (Dothideomycetes). The further newly described Spegazzinia species supported the placement of the genus as a basal clade in Didymosphaeriaceae (Thambugala et al. 2017; Samarakoon et al. 2020a). However, only the asexual morph of Spegazzinia species is determined, their typical conidia originate terminally at the apex of basauxic conidiophores (Mena-Portales et al. 2017), separating them from the further species in the Didymosphaeriaceae with only coelomycetous asexual morphs (Thambugala et al. 2017). The other genus with which Agrorhizomyces was in sister position, Dictyoarthrinium, was introduced in 1952 by Hughes based on D. quadratum (Samarakoon et al. 2020b). Until Vu et al. (2019) sequenced D. sacchari (CBS 529.73), all Dictyoarthrinium species were introduced based on their morphological data. Although Hyde et al. (2020a, b) still accommodated the genus in the Apiosporaceae, based on their basauxic conidiogenous cells that are like those formed by Spegazzinia species, Samarakoon et al. (2020b) placed Dictyoarthrinium in the Didymosphaeriaceae, as well. Agrorhizomyces grouped with two asexual hyphomycete genera in the family that consist of mostly ascomata-forming species and anamorphic taxa with coelomycetous asexual morphs. We found coelomycetous structures, that were globose and sterile without conidia and conidiophores and resembled those formed by Spegazzinia and Dictyoarthrinium. Although each morph was sterile or immature, comprising no spores and other characteristic features, the presence of these globose structures, strengthened the affiliation of A. patris to the Didymosphaeriaceae, although they were never observed in the two neighboring genera.
The ITS BLASTn search (08/01/2024) in GenBank resulted only three unpublished, almost identical (> 99% similarity) environmental uncultured sequences with ~ 93% similarity with our ITS sequence from A. patris. One (MF569188) was obtained from soil from the above-mentioned location (Bloomington-Normal, IL USA; Beck 2017), while the other (KX192630) was derived from soil from Illinois, Champaign County, USA. The third (ON696029) originated from the above listed Australian site (Hassan et al. 2022). Albeit the sampling numbers are small, it seems, the novel DSE taxa (and their close relative) co-occurred at the same sites at their original location and also at North American and Australian locations (Beck 2017; Hassan et al. 2022).
Spegazzinia and Dictyoarthrinium species, are found on a wide range of dead plant material in various habitats, the former also on various host species. Although we have limited information on their lifestyle and function, we are not aware that they are plant pathogenic. Some Spegazzinia species, such as S. tessarthra, have been identified as saprobes and endophytes (Samarakoon et al. 2020a). Most of the Dictyoarthrinium species are considered saprobes showing affinity to monocotyledonous plants. The genus is widely distributed across the tropics, mainly in terrestrial environments (Samarakoon et al. 2020b). We are not aware of that A. patris was isolated from soil, living plant tissues or debris. We found no similar ITS sequences of A. patris in the GenBank and neither in our metabarcode database of natural grassland ecosystem. The dominant presence of A. patris in the roots of wheat in our sampling site was strengthened by the ITS2-based metabarcoding study of the soil fungal community in the same sites and parcels. Each three genera are plant-associated, and endophytic lifestyle cannot be ruled out in the case of Spegazzinia and Dictyoarthrinium species either. Although one of the six A. patris isolates had a slight but significant negative effect on the host biomass, the here reported isolates might be beneficial also in ways other than plant growth promotion (Ruotsalainen et al. 2022).
Our results illustrate also that environments where the taxon diversity of nonpathogenic fungi are less studied, like the soils of agronomic croplands, might be an unexplored reservoir of hidden fungal diversity. Revealing this diversity could help us understanding the spread and functional role of root endophytic fungi and might lead to beneficial agricultural applications.
Data availability
Datasets and specimens and strains and their data were deposited at the Hungarian Natural History Museum, Budapest (BP), Westerdijk Fungal Biodiversity Institute, NCBI GenBank, Figshare repository and MycoBank.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Andrade-Linares DR, Franken P (2013) Fungal endophytes in plant roots: taxonomy, colonization patterns, and functions. In: Aroca R (ed) Symbiotic endophytes. Springer, Berlin Heidelberg, Berlin, pp 311–334
Andrade-Linares DR, Grosch R, Restrepo S, Krumbein A, Franken P (2011) Effects of dark septate endophytes on tomato plant performance. Mycorrhiza 21:413–422. https://doi.org/10.1007/s00572-010-0351-1
Ariyawansa HA, Tanaka K, Thambugala KM, Phookamsak R, Tian Q, Camporesi E, Hongsanan S, Monkai J, Wanasinghe DN, Mapook A, Chukeatirote E, Kang JC, Xu JC, McKenzie EHC, Gareth Jones EBG, Hyde KD (2014) A molecular phylogenetic reappraisal of the Didymosphaeriaceae (= Montagnulaceae). Fung Div 68:69–104. https://doi.org/10.1007/s13225-014-0305-6
Ashrafi S, Knapp DG, Blaudez D, Chalot M, Maciá-Vicente JG, Zagyva I, Dababat AA, Maier W, Kovács GM (2018) Inhabiting plant roots, nematodes, and truffles –Polyphilus, a new helotialean genus with two globally distributed species. Mycologia 110:286–299. https://doi.org/10.1080/00275514.2018.1448167
Ashrafi S, Wennrich JP, Becker Y, Maciá-Vicente JG, Brißke-Rode A, Daub M, Thünen T, Dababat AA, Finckh MR, Stadler M, Maier W (2023) Polydomus karssenii gen. nov. sp. nov. is a dark septate endophyte with a bifunctional lifestyle parasitising eggs of plant parasitic cyst nematodes (Heterodera spp.). IMA Fungus 14:6. https://doi.org/10.1186/s43008-023-00113-w
Beck AM (2017) Invasive Lespedeza cuneata and its relationship to soil microbes and plant-soil feedback. Dissertation University of Illinois
Beschoren da Costa P, Benucci GMN, Chou MY, Van Wyk J, Chretien M, Bonito G (2022) Soil origin and plant genotype modulate switchgrass aboveground productivity and root microbiome assembly. mBio 13:e0007922. https://doi.org/10.1128/mbio.00079-22
Boonmee S, Wanasinghe DN, Calabon MS, Huanraluek N, Chandrasiri SKU, Jones GEB, Rossi W, Leonardi M, Singh SK, Rana S, Singh PN, Maurya DK, Lagashetti AC, Choudhary D, Dai YC, Zhao CL, Mu YH, Yuan HS, He SH, Phookamsak R, Jiang HB, Martín MP, Dueñas M, Telleria MT, Kałucka IL, Jagodziński AM, Liimatainen K, Pereira DS, Phillips AJL, Suwannarach N, Kumla J, Khuna S, Lumyong S, Potter TB, Shivas RG, Sparks AH, Vaghefi N, Abdel-Wahab MA, Abdel-Aziz FA, Li GJ, Lin WF, Singh U, Bhatt RP, Lee HB, Nguyen TTT, Kirk PM, Dutta AK, Acharya K, Sarma VV, Niranjan M, Rajeshkumar KC, Ashtekar N, Lad S, Wijayawardene NN, Bhat DJ, Xu RJ, Wijesinghe SN, Shen HW, Luo ZL, Zhang JY, Sysouphanthong P, Thongklang N, Bao DF, Aluthmuhandiram JVS, Abdollahzadeh J, Javadi A, Dovana F, Usman M, Khalid AN, Dissanayake AJ, Telagathoti A, Probst M, Peintner U, Garrido-Benavent I, Bóna L, Merényi Z, Boros L, Zoltán B, Stielow JB, Jiang N, Tian CM, Shams E, Dehghanizadeh F, Pordel A, Javan-Nikkhah M, Denchev TT, Denchev CM, Kemler M, Begerow D, Deng CY, Harrower E, Bozorov T, Kholmuradova T, Gafforov Y, Abdurazakov A, Xu JC, Mortimer PE, Ren GC, Jeewon R, Maharachchikumbura SSN, Phukhamsakda C, Mapook A, Hyde KD (2021) Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers 111:1–335. https://doi.org/10.1007/s13225-021-00489-3
Calabon MS, Jones EB, Hyde KD, Boonmee S, Tibell S, Tibell L, Pang KL, Phookamsak R (2021) Phylogenetic assessment and taxonomic revision of Halobyssothecium and Lentithecium (Lentitheciaceae, Pleosporales). Mycol Progress 20:701–720. https://doi.org/10.1007/s11557-021-01692-x
Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G (2004) MycoBank: an online initiative to launch mycology into the 21st century. Stud Mycol 50:19–22
Crous PW, Carnegie AJ, Wingfield MJ, Sharma R, Mughini G, Noordeloos ME, Santini A, Shouche YS, Bezerra JDP, Dima B, Guarnaccia V, Imrefi I, Jurjević Ž, Knapp DG, Kovács GM, Magistà D, Perrone G, Rämä T, Rebriev YA, Shivas RG, Singh SM, Souza-Motta CM, Thangavel R, Adhapure NN, Alexandrova AV, Alfenas AC, Alfenas RF, Alvarado P, Alves AL, Andrade DA, Andrade JP, Barbosa RN, Barili A, Barnes CW, Baseia IG, Bellanger JM, Berlanas C, Bessette AE, Bessette AR, Biketova AY, Bomfim FS, Brandrud TE, Bransgrove K, Brito ACQ, Cano-Lira JF, Cantillo T, Cavalcanti AD, Cheewangkoon R, Chikowski RS, Conforto C, Cordeiro TRL, Craine JD, Cruz R, Damm U, de Oliveira RJV, de Souza JT, de Souza HG, Dearnaley JDW, Dimitrov RA, Dovana F, Erhard A, Esteve-Raventós F, Félix CR, Ferisin G, Fernandes RA, Ferreira RJ, Ferro LO, Figueiredo CN, Frank JL, Freire KTLS, García D, Gené J, Gêsiorska A, Gibertoni TB, Gondra RAG, Gouliamova DE, Gramaje D, Guard F, Gusmão LFP, Haitook S, Hirooka Y, Houbraken J, Hubka V, Inamdar A, Iturriaga T, Iturrieta-González I, Jadan M, Jiang N, Justo A, Kachalkin AV, Kapitonov VI, Karadelev M, Karakehian J, Kasuya T, Kautmanová I, Kruse J, Kušan I, Kuznetsova TA, Landell MF, Larsson KH, Lee HB, Lima DX, Lira CRS, Machado AR, Madrid H, Magalhães OMC, Majerova H, Malysheva EF, Mapperson RR, Marbach PAS, Martín MP, Martín-Sanz A, Matočec N, McTaggart AR, Mello JF, Melo RFR, Mešić A, Michereff SJ, Miller AN, Minoshima A, Molinero-Ruiz L, Morozova OV, Mosoh D, Nabe M, Naik R, Nara K, Nascimento SS, Neves RP, Olariaga I, Oliveira RL, Oliveira TGL, Ono T, Ordoñez ME, Ottoni AM, Paiva LM, Pancorbo F, Pant B, Pawłowska J, Peterson SW, Raudabaugh DB, Rodríguez-Andrade E, Rubio E, Rusevska K, Santiago ALCMA, Santos ACS, Santos C, Sazanova NA, Shah S, Sharma J, Silva BDB, Siquier JL, Sonawane MS, Stchigel AM, Svetasheva T, Tamakeaw N, Telleria MT, Tiago PV, Tian CM, Tkalčec Z, Tomashevskaya MA, Truong HH, Vecherskii MV, Visagie CM, Vizzini A, Yilmaz N, Zmitrovich IV, Zvyagina EA, Boekhout T, Kehlet T, Læssøe T, Groenewald JZ (2019) Fungal Planet description sheets: 868–950. Persoonia 42:291–473. https://doi.org/10.3767/persoonia.2019.42.11
Crous PW, Wingfield MJ, Chooi YH, Gilchrist CLM, Lacey E, Pitt JI, Roets F, Swart WJ, Cano-Lira JF, Valenzuela-Lopez N, Hubka V, Shivas RG, Stchigel AM, Holdom DG, Jurjević Ž, Kachalkin AV, Lebel T, Lock C, Martín MP, Tan YP, Tomashevskaya MA, Vitelli JS, Baseia IG, Bhatt VK, Brandrud TE, De Souza JT, Dima B, Lacey HJ, Lombard L, Johnston PR, Morte A, Papp V, Rodríguez A, Rodríguez-Andrade E, Semwal KC, Tegart L, Abad ZG, Akulov A, Alvarado P, Alves A, Andrade JP, Arenas F, Asenjo C, Ballarà J, Barrett MD, Berná LM, Berraf-Tebbal A, Bianchinotti MV, Bransgrove K, Burgess TI, Carmo FS, Chávez R, Čmoková A, Dearnaley JDW, de Santiago ALCM, Freitas-Neto JF, Denman S, Douglas B, Dovana F, Eichmeier A, Esteve-Raventós F, Farid A, Fedosova AG, Ferisin G, Ferreira RJ, Ferrer A, Figueiredo CN, Figueiredo YF, Reinoso-Fuentealba CG, Garrido-Benavent I, Cañete-Gibas CF, Gil-Durán C, Glushakova AM, Gonçalves MFM, González M, Gorczak M, Gorton C, Guard FE, Guarnizo AL, Guarro J, Gutiérrez M, Hamal P, Hien LT, Hocking AD, Houbraken J, Hunter GC, Inácio CA, Jourdan M, Kapitonov VI, Kelly L, Khanh TN, Kisło K, Kiss L, Kiyashko A, Kolařík M, Kruse J, Kubátová A, Kučera V, Kučerová I, Kušan I, Lee HB, Levicán G, Lewis A, Liem NV, Liimatainen K, Lim HJ, Lyons MN, Maciá-Vicente JG, Magaña-Dueñas V, Mahiques R, Malysheva EF, Marbach PAS, Marinho P, Matočec N, McTaggart AR, Mešić A, Morin L, Muñoz-Mohedano JM, Navarro-Ródenas A, Nicolli CP, Oliveira RL, Otsing E, Ovrebo CL, Pankratov TA, Paños A, Paz-Conde A, Pérez-Sierra A, Phosri C, Pintos Á, Pošta A, Prencipe S, Rubio E, Saitta A, Sales LS, Sanhueza L, Shuttleworth LA, Smith J, Smith ME, Spadaro D, Spetik M, Sochor M, Sochorová Z, Sousa JO, Suwannasai N, Tedersoo L, Thanh HM, Thao LD, Tkalčec Z, Vaghefi N, Venzhik AS, Verbeken A, Vizzini A, Voyron S, Wainhouse M, Whalley AJS, Wrzosek M, Zapata M, Zeil-Rolfe I, Groenewald JZ (2020) Fungal Planet description sheets: 1042–1111. Persoonia 44:301–459. https://doi.org/10.3767/persoonia.2020.44.11
Crous PW, Begoude BAD, Boers J, Braun U, Declercq B, Dijksterhuis J, Elliott TF, Garay-Rodriguez GA, Jurjević Ž, Kruse J, Linde CC, Loyd A, Mound L, Osieck ER, Rivera-Vargas LI, Quimbita AM, Rodas CA, Roux J, Schumacher RK, StarinkWillemse M, Thangavel R, Trappe JM, van Iperen AL, Van Steenwinkel C, Wells A, Wingfield MJ, Yilmaz N, Groenewald JZ (2022) New and Interesting Fungi. Fungal Syst Evol 10:19–90. https://doi.org/10.3114/fuse.2022.10.02
Crous PW, Cowan DA, Maggs-Kölling G, Yilmaz N, Thangavel R, Wingfield MJ, Noordeloos ME, Dima B, Brandrud TE, Jansen GM, Morozova OV, Vila J, Shivas RG, Tan YP, Bishop-Hurley S, Lacey E, Marney TS, Larsson E, Le Floch G, Lombard L, Nodet P, Hubka V, Alvarado P, Berraf-Tebbal A, Reyes JD, Delgado G, Eichmeier A, Jordal JB, Kachalkin AV, Kubátová A, Maciá-Vicente JG, Malysheva EF, Papp V, Rajeshkumar KC, Sharma A, Spetik M, Szabóová D, Tomashevskaya MA, Abad JA, Abad ZG, Alexandrova AV, Anand G, Arenas F, Ashtekar N, Balashov S, Bañares Á, Baroncelli R, Bera I, Biketova AY, Blomquist CL, Boekhout T, Boertmann D, Bulyonkova TM, Burgess TI, Carnegie AJ, Cobo-Diaz JF, Corriol G, Cunnington JH, da Cruz MO, Damm U, Davoodian N, de A Santiago ALCM, Dearnaley J, de Freitas LWS, Dhileepan K, Dimitrov R, Di Piazza S, Fatima S, Fuljer F, Galera H, Ghosh A, Giraldo A, Glushakova AM, Gorczak M, Gouliamova DE, Gramaje D, Groenewald M, Gunsch CK, Gutiérrez A, Holdom D, Houbraken J, Ismailov AB, Istel Ł, Iturriaga T, Jeppson M, Jurjević Ž, Kalinina LB, Kapitonov VI, Kautmanová I, Khalid AN, Kiran M, Kiss L, Kovács Á, Kurose D, Kušan I, Lad S, Læssøe T, Lee HB, Luangsa-Ard JJ, Lynch M, Mahamedi AE, Malysheva VF, Mateos A, Matočec N, Mešić A, Miller AN, Mongkolsamrit S, Moreno G, Morte A, Mostowfizadeh-Ghalamfarsa R, Naseer A, Navarro-Ródenas A, Nguyen TTT, Noisripoom W, Ntandu JE, Nuytinck J, Ostrý V, Pankratov TA, Pawłowska J, Pecenka J, Pham THG, Polhorský A, Pošta A, Raudabaugh DB, Reschke K, Rodríguez A, Romero M, Rooney-Latham S, Roux J, Sandoval-Denis M, Smith MT, Steinrucken TV, Svetasheva TY, Tkalčec Z, van der Linde EJ, V D Vegte M, Vauras J, Verbeken A, Visagie CM, Vitelli JS, Volobuev SV, Weill A, Wrzosek M, Zmitrovich IV, Zvyagina EA, Groenewald JZ (2021) Fungal Planet description sheets: 1182–1283. Persoonia 46:313–528. https://doi.org/10.3767/persoonia.2021.46.11
Fors RO, Patreze CM, Louro Berbara RL, Carbone Carneiro MA, Saggin-Júnior OJ (2020) Dark septate endophytic fungi associated with sugarcane plants cultivated in São Paulo. Brazil Diversity 12:351. https://doi.org/10.3390/d12090351
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol Ecol 2:13–118. https://doi.org/10.1111/j.1365-294x.1993.tb00005.x
Gdanetz K, Trail F (2017) The wheat microbiome under four management strategies, and potential for endophytes in disease protection. Phytobiomes 1:158–168. https://doi.org/10.1094/PBIOMES-05-17-0023-R
Gooding GV, Lucas GB (1959) Factors influencing sporangial formation and zoospore activity in Phytophthora parasitica var. nicotianae. Phytopathol 49:277–281
Grünig CR, Queloz V, Sieber TN (2011) Structure of diversity in dark septate endophytes: from species to genes. In: Pirttilä AM, Frank AC (eds) Endophytes of forest trees: biology and applications. Springer, Dordrecht, pp 3–30
Hall G (1986) Demonstration of chlamydospores and evidence for microsclerotia in Periconia macrospinosa. Trans Br Mycol Soc 86:347–349. https://doi.org/10.1016/S0007-1536(86)80226-1
Hassan K, Carrillo Y, Nielsen UN (2022) Prolonged drought causes negative plant-soil feedbacks in grassland species under field conditions. Soil Biol Biochem 172:108772. https://doi.org/10.1016/j.soilbio.2022.108772
Hughes SJ (1952) Fungi from the Gold Coast. 11. Mycol Pap 50:1–104
Hyde KD, Norphanphoun C, Abreu VP, Bazzicalupo A, Chethana KWT, Clericuzio M, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, He MQ, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena RS, Karunarathna A, Konta S, Kušan I, Lee H, Li J, Lin CG, Liu NG, Lu YZ, Luo ZL, Manawasinghe IS, Mapook A, Perera RH, Phookamsak R, Phukhamsakda C, Siedlecki I, Soares AM, Tennakoon DS, Tian Q, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Abdel-Aziz FA, Li WJ, Senanayake IC, Shang QJ, Daranagama DA, de Silva NI, Thambugala KM, Abdel-Wahab MA, Bahkali AH, Berbee ML, Boonmee S, Bhat DJ, Bulgakov TS, Buyck B, Camporesi E, Castañeda-Ruiz RF, Chomnunti P, Doilom M, Dovana F, Gibertoni TB, Jadan M, Jeewon R, Jones EBG, Kang JC, Karunarathna SC, Lim YW, Liu JK, Liu ZY, Plautz HL Jr, Lumyong S, Maharachchikumbura SSN, Matočec N, McKenzie EHC, Mešić A, Miller D, Pawłowska J, Pereira OL, Promputtha I, Romero AI, Ryvarden L, Su HY, Suetrong S, Tkalčec Z, Vizzini A, Wen TC, Wisitrassameewong K, Wrzosek M, Xu JC, Zhao Q, Zhao RL, Mortimer PE, Hyde KD, Norphanphoun C, Abreu VP et al (2017) Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fung Div 87:1–235. https://doi.org/10.1007/s13225-017-0391-3
Hyde KD, Chaiwan N, Norphanphoun C, Boonmee S, Camporesi E, Chethana KWT, Dayarathne MC, de Silva NI, Dissanayake AJ, Ekanayaka AH, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena RS, Jiang HB, Karunarathna A, Lin CG, Liu JK, Liu NG, Lu YZ, Luo ZL, Maharachchimbura SSN, Manawasinghe IS, Pem D, Perera RH, Phukhamsakda C, Samarakoon MC, Senwanna C, Shang QJ, Tennakoon DS, Thambugala KM, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Zhang JF, Zhang SN, Bulgakov TS, Bhat DJ, Cheewangkoon R, Goh TK, Jones EBG, Kang JC, Jeewon R, Liu ZY, Lumyong S, Kuo CH, McKenzie EHC, Wen TC, Yan JY, Zhao Q (2018) Mycosphere notes 169–224. Mycosphere 9:271–430. https://doi.org/10.5943/mycosphere/9/2/8
Hyde KD, Jeewon R, Chen YJ, Bhunjun CS, Calabon MS, Jiang HB, Lin CG, Norphanphoun C, Sysouphanthong P, Pem D, Tibpromma S, Zhang Q, Doilom M, Jayawardena RS, Liu JK, Maharachchikumbura SNS, Phukhamsakda C, Phookamsak R, Al-Sadi AM, Thongklang N, Wang Y, Gafforov Y, Jones EBG, Lumyong S (2020a) The numbers of fungi: is the descriptive curve flattening? Fung Div 103:219–271. https://doi.org/10.1007/s13225-020-00458-2
Hyde KD, Norphanphoun C, Maharachchikumbura SSN, Bhat DJ, Jones EBG, Bundhun D, Chen YJ, Bao DF, Boonmee S, Calabon MS, Chaiwan N, Chethana KWT, Dai DQ, Dayarathne MC, Devadatha B, Dissanayake AJ, Dissanayake LS, Doilom M, Dong W, Fan XL, Goonasekara ID, Hongsanan S, Huang SK, Jayawardena RS, Jeewon R, Karunarathna A, Konta S, Kumar V, Lin CG, Liu JK, Liu NG, Luangsa-ard J, Lumyong S, Luo ZL, Marasinghe DS, McKenzie EHC, Niego AGT, Niranjan M, Perera RH, Phukhamsakda C, Rathnayaka AR, Samarakoon MC, Samarakoon SMBC, Sarma VV, Senanayake IC, Shang QJ, Stadler M, Tibpromma S, Wanasinghe DN, Wei DP, Wijayawardene NN, Xiao YP, Yang J, Zeng XY, Zhang SN, Xiang MM (2020b) Refined Families of Sordariomycetes. Mycosphere 11:305–1059. https://doi.org/10.5943/mycosphere/9/2/8
Hyde KD, Suwannarach N, Jayawardena RS, Manawasinghe IS, Liao CF, Doilom M, Cai L, Zhao P, Buyck B, Phukhamsakda C, Su WX, Fu YP, Li Y, Zhao RL, He MQ, Li JX, Tibpromma S, Lu L, Tang X, Kang JC, Ren GC, Gui H, Hofstetter V, Ryoo R, Antonín V, Hurdeal VG, Gentekaki E, Zhang JY, Lu YZ, Senanayake IC, Yu FM, Zhao Q, Bao DF (2021) Mycosphere notes 325–344 – Novel species and records of fungal taxa from around the world. Mycosphere 12:1101–1156. https://doi.org/10.5943/mycosphere/12/1/14
Jumpponen A, Herrera J, Porras-Alfaro A, Rudgers J (2017) Biogeography of root-associated fungal endophytes. In: Tedersoo L (ed) Biogeography of Mycorrhizal Symbiosis Springer, Dordrecht, pp 195–222
Jumpponen A, Trappe JM (1998) Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi. New Phytol 140:295–310. https://doi.org/10.1046/j.1469-8137.1998.00265.x
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Knapp DG, Kovács GM (2016) Interspecific metabolic diversity of root-colonizing endophytic fungi revealed by enzyme activity tests. FEMS Microbiol Ecol 92:fiw90
Knapp DG, Pintye A, Kovács GM (2012) The dark side is not fastidious – dark septate endophytic fungi of native and invasive plants of semiarid sandy areas. PLoS ONE 7:e32570
Knapp DG, Kovács GM, Zajta E, Groenewald JZ, Crous PW (2015) Dark septate endophytic pleosporalean genera from semiarid areas. Persoonia 35:87–100. https://doi.org/10.3767/003158515X688433
Knapp DG, Németh JB, Barry K, Hainaut M, Henrissat B, Johnson J, Kuo A, Lim JHP, Lipzen A, Nolan M, Ohm RA, Tamás L, Grigoriev IV, Spatafora JW, Nagy LG, Kovács GM (2018) Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci Rep 8:6321. https://doi.org/10.1038/s41598-018-24686-4
Knapp DG, Imrefi I, Boldpurev E, Csíkos S, Akhmetova G, Berek-Nagy PJ, Otgonsuren B, Kovács GM (2019) Root-colonizing endophytic fungi of the dominant grass Stipa krylovii from a Mongolian steppe grassland. Front Microbiol 10:2565. https://doi.org/10.3389/fmicb.2019.02565
Knapp DG, Akhmetova GK, Kovács GM, Dababat AA, Maier W, Ashrafi S (2022) Two new root endophyte and nematode cyst parasite species of the widely distributed genus Laburnicola. Mycol Prog 21:99. https://doi.org/10.1007/s11557-022-01725-y
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lefebvre CL, Johnson AG, Sherwin HS (1949) An undescribed species of Periconia. Mycologia 41:416–419. https://doi.org/10.1080/00275514.1949.12017787
Liu ZP, Zhang SN, Cheewangkoon R, Zhao Q, Liu JK (2022) Crassoascoma gen. nov. (Lentitheciaceae, Pleosporales): Unrevealing microfungi from the Qinghai-Tibet Plateau in China. Diversity 14:15. https://doi.org/10.3390/d14010015
Luo J, Walsh E, Naik A, Zhuang W, Zhang K, Cai L, Zhang N (2014) Temperate pine barrens and tropical rain forests are both rich in undescribed fungi. PLoS ONE 9:e103753. https://doi.org/10.1371/journal.pone.0103753
Luo ZL, Bahkali AH, Liu XY, Phookamsak R, Zhao YC, Zhou DQ, Su HY, Hyde KD (2016) Poaceascoma aquaticum sp. nov. (Lentitheciaceae), a new species from submerged bamboo in freshwater. Phytotaxa 253:71–80. https://doi.org/10.11646/phytotaxa.253.1.5
Maciá-Vicente JG, Shi YN, Cheikh-Ali Z, Grün P, Glynou K, Kia SH, Piepenbring M, Bode HB (2018) Metabolomics-based chemotaxonomy of root endophytic fungi for natural products discovery. Environ Microbiol 20:1253–1270. https://doi.org/10.1111/1462-2920.14072
Mandyam K, Jumpponen A (2005) Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud Mycol 53:173–189. https://doi.org/10.3114/sim.53.1.173
Mandyam K, Loughin T, Jumpponen A (2010) Isolation and morphological and metabolic characterization of common endophytes in annually burned tallgrass prairie. Mycologia 102:813–821. https://doi.org/10.3852/09-212
Mayer Z, Sasvári Z, Szentpéteri V, Pethőné Rétháti B, Vajna B, Posta K (2019) Effect of long-term cropping systems on the diversity of the soil bacterial communities. Agron 9:878. https://doi.org/10.3390/agronomy9120878
Megyes M, Borsodi AK, Árendás T, Márialigeti K (2021) Variations in the diversity of soil bacterial and archaeal communities in response to different long-term fertilization regimes in maize fields. Appl Soil Ecol 168:104120. https://doi.org/10.1016/j.apsoil.2021.104120
Mena-Portales J, Cantillo-Perez T, Minter DW (2017) A new species of the conidial fungal genus Spegazzinia (Pleosporales, Didymosphaeriaceae) collected on sugarcane in Cuba. Phytotaxa 331:295–298. https://doi.org/10.11646/phytotaxa.331.2.14
Németh JB, Knapp DG, Kósa A, Hegedűs PÁ, Herczeg G, Vági P, Kovács GM (2022) Micro-scale experimental system coupled with fluorescence-based estimation of fungal biomass to study utilisation of plant substrates. Microb Ecol 83:714–723. https://doi.org/10.1007/s00248-021-01794-9
Newsham KK (2011) A meta-analysis of plant responses to dark septate root endophytes. New Phytol 190:783–793. https://doi.org/10.1111/j.1469-8137.2010.03611.x
Pereira E, Vázquez de Aldana BR, San Emeterio L, Zabalgogeazcoa I (2019) A survey of culturable fungal endophytes from Festuca rubra subsp. pruinosa, a grass from marine cliffs, reveals a core microbiome. Front Microbiol 16:3321. https://doi.org/10.3389/fmicb.2018.03321
Petrini O (1991) Fungal endophytes of tree leaves. In: Andrews JH, Hirano SH (eds) Microbial ecology of leaves. Springer, New York, New York, pp 179–197
Phookamsak R, Manamgoda DS, Li WJ, Dai DQ, Singtripop C, Hyde KD (2015) Poaceascoma helicoides gen et sp. nov., a new genus with scolecospores in Lentitheciaceae. Cryptogam Mycol 36:225–236. https://doi.org/10.7872/crym/v36.iss2.2015.225
Pintye A, Knapp DG (2021) Two pleosporalean root-colonizing fungi, Fuscosphaeria hungarica gen. et sp. nov. and Delitschia chaetomioides, from a semiarid grassland in Hungary. Mycol Prog 20:39–50. https://doi.org/10.1007/s11557-020-01655-8
Porras-Alfaro A, Bayman P (2011) Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol 49:291–315. https://doi.org/10.1146/annurev-phyto-080508-081831
Rajeshkumar KC, Varma RK, Sruthi OP, Gautam AK, Crous PW (2023) Groenewaldia (Lentitheciaceae), a new corticolous fungal genus from India. Mycol Prog 22:43. https://doi.org/10.1007/s11557-023-01888-3
Raza M, Zhang ZF, Hyde KD, Diao YZ, Cai L (2019) Culturable plant pathogenic fungi associated with sugarcane in southern China. Fung Div 99:1–104. https://doi.org/10.1007/s13225-019-00434-5
Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98. https://doi.org/10.3852/mycologia.97.1.84
Rehner SA, Samuels GS (1994) Taxonomy and phylogeny of Gliocladium analysed for nuclear large subunit ribosomal DNA sequences. Mycol Res 98:625–634
Ren G, Wanasinghe DN, de Farias ARG, Hyde KD, Yasanthika E, Xu J, Balasuriya A, Chethana KWT, Gui H (2022) Taxonomic Novelties of Woody Litter Fungi (Didymosphaeriaceae, Pleosporales) from the Greater Mekong Subregion. Biology 11:1660. https://doi.org/10.3390/biology11111660
Rodriguez RJ, White JF Jr, Arnold AE, Redman ARA (2009) Fungal endophytes: diversity and functional roles. New Phytol 182:314–330. https://doi.org/10.1111/j.1469-8137.2009.02773.x
Romero-Jiménez MJ, Rudgers JA, Jumpponen A, Herrera J, Hutchinson M, Kuske C, Dunbar J, Knapp DG, Kovács GM, Porras-Alfaro A (2022) Darksidea phi, sp. nov., a dark septate root-associated fungus in foundation grasses in North American Great Plains. Mycologia 114:254–269. https://doi.org/10.1080/00275514.2022.2031780
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Rudgers JA, Fox S, Porras-Alfaro A, Herrera J, Reazin C, Kent DR, Souza L, Chung A, Jumpponen A (2022) Biogeography of root-associated fungi in foundation grasses of North American Plains. J Biogeogr 49:22–37. https://doi.org/10.1111/jbi.14260
Ruotsalainen AL, Kauppinen M, Wäli PR, Saikkonen K, Helander M, Tuomi J (2022) Dark septate endophytes: mutualism from by-products? Trends Plant Sci 27:247–254. https://doi.org/10.1016/j.tplants.2021.10.001
Samarakoon BC, Phookamsak R, Wanasinghe DN, Chomnunti P, Hyde KD, McKenzie EHC, Promputtha I, Xu JC, Li YJ (2020a) Taxonomy and phylogenetic appraisal of Spegazzinia musae sp. nov. and S. deightonii (Didymosphaeriaceae, Pleosporales) on Musaceae from Thailand. MycoKeys 70:19–37. https://doi.org/10.3897/mycokeys.70.52043
Samarakoon BC, Wanasinghe DN, Samarakoon MC, Phookamsak R, McKenzie EHC, Chomnunti P, Hyde KD, Lumyong S, Karunarathna SC (2020b) Multi-gene phylogenetic evidence suggests Dictyoarthrinium belongs in Didymosphaeriaceae (Pleosporales, Dothideomycetes) and Dictyoarthrinium musae sp. nov. on Musa from Thailand. MycoKeys 71:101–118. https://doi.org/10.3897/mycokeys.71.55493
Sieber TN, Grünig CR (2013) Fungal root endophytes. In: Waisel Y, Eshel A, Beeckman T, Kafkafi U (ed) Plant Roots: the Hidden Half. Marcel Dekker, New York
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337. https://doi.org/10.1007/s13127-011-0056-0
Staden R, Beal KF, Bonfield JK (2000) The Staden package, 1998. Methods Mol Biol 132:115–130. https://doi.org/10.1385/1-59259-192-2:115
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Tan YP, Marney TS, Bishop-Hurley S, Bransgrove KL, Shivas RG (2021) Index Fungorum no. 490. https://www.indexfungorum.org/Publications/Index%20Fungorum%20no.490.pdf. Accessed 17 Mar 2024
Tanaka K, Hirayama K, Yonezawa H, Sato G, Toriyabe A, Kudo H, Hashimoto A, Matsumura M, Harada Y, Kurihara Y, Shirouzu T, Hosoya T (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud Mycol 82:75–136. https://doi.org/10.1016/j.simyco.2015.10.002
Thambugala KM, Wanasinghe DN, Phillips AJL, Camporesi E, Bulgakov TS, Phukhamsakda C, Ariyawansa HA, Goonasekara ID, Phookamsak R, Dissanayake A, Tennakoon DS, Tibpromma S, Chen YY, Liu ZY, Hyde KD (2017) Mycosphere notes 1–50: grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 8:697–796. https://doi.org/10.5943/mycosphere/8/4/13
Ujvári G, Borsodi AK, Megyes M, Mucsi M, Szili-Kovács T, Szabó A, Szalai Z, Jakab G, Márialigeti K (2020) Comparison of soil bacterial communities from juvenile maize plants of a long-term monoculture and a natural grassland. Agronomy 10:341. https://doi.org/10.3390/agronomy10030341
Vajna B, Knapp GD, Dima B, Szalai Z, Kröel-Dulay G, Kovács MG (2021) Effect of a climate manipulation on soil microbial communities in a sandy grassland. In: Szabó D (ed) Acta Microbiologica et Immunologica Hungarica. Volume 68, Akadémia Kiadó, Budapest, pp 125
Valenzuela-Lopez N, Cano-Lira JF, Guarro J, Sutton DA, Wiederhold N, Crous PW, Stchigel AM (2018) Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud Mycol 90:1–69. https://doi.org/10.1016/j.simyco.2017.11.003
Vandenkoornhuyse P, Baldauf SL, Leyval C, Straczek J, Young JPW (2002) Extensive fungal diversity in plant roots. Science 29:2051–2051. https://doi.org/10.1126/science.295.5562.2051
Verkley G, Dukik K, Renfurm R, Göker M, Stielow J (2014) Novel genera and species of coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 32:25–51. https://doi.org/10.3767/003158514X679191
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246
Vohník M, Borovec O, Kolaříková Z, Sudová R, Réblová M (2019) Extensive sampling and high-throughput sequencing reveal Posidoniomyces atricolor gen. et sp. nov. (Aigialaceae, Pleosporales) as the dominant root mycobiont of the dominant Mediterranean seagrass Posidonia oceanica. MycoKeys 55:59. https://doi.org/10.3897/mycokeys.55.35682
Vu D, Groenewald M, De Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud Mycol 92:135–154. https://doi.org/10.1016/j.simyco.2018.05.001
Waldrop MP, Zak DR, Blackwood CB, Curtis CD, Tilman D (2006) Resource availability controls fungal diversity across a plant diversity gradient. Ecol Lett 9:1127–1135. https://doi.org/10.1111/j.1461-0248.2006.00965.x
Wanasinghe DN, Jones EBG, Camporesi E, Dissanayake AJ, Kamolhan S, Mortimer PE, Xu J, Elsalam KA, Hyde KD (2016) Taxonomy and phylogeny of Laburnicola gen. nov. and Paramassariosphaeria gen. nov. (Didymosphaeriaceae, Massarineae, Pleosporales). Fungal Biol 120:1354–1373. https://doi.org/10.1016/j.funbio.2016.06.006
White TJ, BrunsT LS, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols: A Guide to Methods and Applications. Academic Press Inc, New York, pp 315–322
Wijayawardene NN, Crous PW, Kirk PM, Hawksworth DL, Boonmee S, Braun U, Dai DQ, D’souza MJ, Diederich P, Dissanayake A, Doilom M, Hongsanan S, Jones EB, Groenewald JZ, Jayawardena R, Lawrey JD, Liu JK, Lücking R, Madrid H, Manamgoda DS, Muggia L, Nelsen MP, Phookamsak R, Suetrong S, Tanaka K, Thambugala KM, Wanasinghe DN, Wikee S, Zhang Y, Aptroot A, Ariyawansa HA, Bahkali AH, Bhat DJ, Gueidan C, Chomnunti P, De Hoog GS, Knudsen K, Li WJ, McKenzie EH, Miller AN, Phillips AJ, Piątek M, Raja HA, Shivas RS, Slippers B, Taylor JE, Tian Q, Wang Y, Woudenberg JH, Cai L, Jaklitsch WM, Hyde KD (2014) Naming and outline of Dothideomycetes-2014 including proposals for the protection or suppression of generic names. Fungal Divers 69:1–55. https://doi.org/10.1007/s13225-014-0309-2
Wijayawardene NN, Hyde KD, Dai DQ, Sánchez-García M, Goto BT, Saxena RK, Erdoğdu M, Selçuk F, Rajeshkumar KC, Aptroot A, Błaszkowski J, Boonyuen N, da Silva GA, de Souza FA, Dong W, Ertz D, Haelewaters D, Jones EBG, Karunarathna SC, Kirk PM, Kukwa M, Kumla J, Leontyev DV, Lumbsch HT, Maharachchikumbura SSN, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro JS, Oehl F, Pawłowska J, Pem D, Pfliegler WP, Phillips AJL, Pošta A, He MQ, Li JX, Raza M, Sruthi OP, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev YS, Wanasinghe DN, Wijesundara DSA, Wimalaseana SDMK, Madrid H, Zhang GQ, Gao Y, Sánchez-Castro I, Tang LZ, Stadler M, Yurkov A, Thines M (2022) Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13:53–453. https://doi.org/10.5943/mycosphere/13/1/2
Wu X, Hu H, Li S, Zhao J, Li J, Zhang G, Li G, Xiu W (2022) Chemical fertilizer reduction with organic material amendments alters co-occurrence network patterns of bacterium-fungus-nematode communities under the wheat–maize rotation regime. Plant Soil 473:605–623. https://doi.org/10.1007/s11104-022-05314-7
Yang Y, Zhang SN, Yu XD, Liu JK (2022) Pseudokeissleriella bambusicola gen et sp nov (Lentitheciaceae, Pleosporales) from bamboos in Sichuan province China. Phytotaxa. 560:263–273. https://doi.org/10.11646/phytotaxa.560.3.1
Yuan Z, Druzhinina IS, Wang X, Zhang X, Peng L, Labbé J (2020) Insight into a highly polymorphic endophyte isolated from the roots of the halophytic seepweed Suaeda salsa: Laburnicola rhizohalophila sp. nov. (Didymosphaeriaceae, Pleosporales). Fungal Biol 124:327–337. https://doi.org/10.1016/j.funbio.2019.10.001
Zhang Y, Schoch CL, Fournier J, Crous PW, de Gruyter J, Woudenberg JHC, Hirayama K, Tanaka K, Pointing SB, Spatafora JW, Hyde KD (2009) Multi-locus phylogeny of Pleosporales: A taxonomic, ecological and evolutionary re-evaluation. Stud Mycol 64:85–102. https://doi.org/10.3114/sim.2009.64.04
Zhang Y, Crous P, Schoch C, Hyde K (2012) Pleosporales. Fungal Divers 53:1–221. https://doi.org/10.1007/s13225-011-0117-x
Acknowledgements
We are thankful for Dr. Shaun R. Pennycook’s (Manaaki Whenua Landcare Research, New Zeland) help in nomenclatural questions, for Dr. Anthony Yannarell’s (University of Illinois at Urbana-Champaign, USA) help with information on environmental sequences, for Dr. Balázs Vajna’s (Eötvös Loránd University, Hungary) help in screening of our fungal metabarcode databases, for Dr. Károly Bóka’s and Dr. Zoltán Kristóf’s (Eötvös Loránd University, Hungary) help in microscopy. Anonymous reviewers and section editor Dr. Hans-Josef Schroers (Agricultural Institute of Slovenia, Slovenia) provided valuable comments on the manuscript.
Funding
Open access funding provided by Eötvös Loránd University. The work was supported by the National Research, Development and Innovation Office of Hungary (projects: K139026; Institutional Excellence Program 2020 (TKP2020-IKA-05), Diagnostics and Therapy 2; GINOP-2.3.2–15-2016–00056.)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Section Editor: Hans-Josef Schroers
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Online Resource 1.
Visualization of hyphae colonizing wheat roots. a: Poaceascoma zborayi strain BDT15 (CBS 151097), colonizing wheat roots with dark septate hyphae, stained with aniline blue b: BDT15 (CBS 151097), colonizing wheat roots with dark septate hyphae and a microsclerotia c: Agrorhizomyces patris strain BBT01 (CBS 151043), microsclerotia in wheat root, stained with WGA-Alexafluor®488 d: same strain colonizing wheat root, stained with aniline blue. Scale bars: 100 μm (JPG 921 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Imrefi, I., Knapp, D.G. & Kovács, G.M. Poaceascoma zborayi sp. nov. and Agrorhizomyces patris gen. et spec. nov. – Two novel dark septate endophytes colonizing wheat (Triticum aestivum) roots from a cropland in Hungary. Mycol Progress 23, 35 (2024). https://doi.org/10.1007/s11557-024-01970-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-024-01970-4