Abstract
Purpose
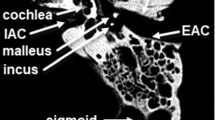
Computational surgical planning tools could help develop novel skull base surgical approaches that improve safety and patient outcomes. This defines a need for automated skull base segmentation to improve the usability of surgical planning software. The objective of this work was to design and validate an algorithm for atlas-based automated segmentation of skull base structures in individual image sets for skull base surgical planning.
Methods
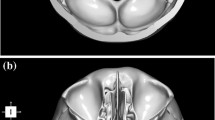
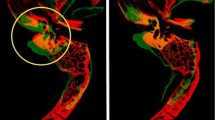
Advanced Normalization Tools software was used to construct a synthetic CT template from 6 subjects, and skull base structures were manually segmented to create a reference atlas. Landmark registration followed by Elastix deformable registration was applied to the template to register it to each of the 30 trusted reference image sets. Dice coefficient, average Hausdorff distance, and clinical usability scoring were used to compare the atlas segmentations to those of the trusted reference image sets.
Results
The mean for average Hausdorff distance for all structures was less than 2 mm (mean for 95th percentile Hausdorff distance was less than 5 mm). For structures greater than 2.5 mL in volume, the average Dice coefficient was 0.73 (range 0.59–0.82), and for structures less than 2.5 mL in volume the Dice coefficient was less than 0.7. The usability scoring survey was completed by three experts, and all structures met the criteria for acceptable effort except for the foramen spinosum, rotundum, and carotid artery, which required more than minor corrections.
Conclusion
Currently available open-source algorithms, such as the Elastix deformable algorithm, can be used for automated atlas-based segmentation of skull base structures with acceptable clinical accuracy and minimal corrections with the use of the proposed atlas. The first publicly available CT template and anterior skull base segmentation atlas being released (available at this link: http://hdl.handle.net/1773/46259) with this paper will allow for general use of automated atlas-based segmentation of the skull base.




Similar content being viewed by others
Availability of data and material
Synthetic CT template and segmentation file are available for download http://hdl.handle.net/1773/46259.
References
Schwartz TH, Morgenstern PF, Anand VK (2019) Lessons learned in the evolution of endoscopic skull base surgery. J Neurosurg 130(2):337–346. https://doi.org/10.3171/2018.10.JNS182154
Tham T, Costantino P, Bruni M, Langer D, Boockvar J, Singh P (2015) Multiportal combined transorbital and transnasal endoscopic resection of fibrous dysplasia. J Neurol Surg Rep 76(2):e291-296. https://doi.org/10.1055/s-0035-1566126
Zhang X, Tabani H, El-Sayed I, Meybodi AT, Griswold D, Mummaneni P, Benet A (2016) Combined endoscopic transoral and endonasal approach to the jugular foramen: a multiportal expanded access to the clivus. World Neurosurg 95:62–70. https://doi.org/10.1016/j.wneu.2016.07.073
Schwartz TH, Fraser JF, Brown S, Tabaee A, Kacker A, Anand VK (2008) Endoscopic cranial base surgery: classification of operative approaches. Neurosurgery 62(5):991–1002; discussion 1002–1005. https://doi.org/10.1227/01.neu.0000325861.06832.06
Liu JK, Decker D, Schaefer SD, Moscatello AL, Orlandi RR, Weiss MH, Couldwell WT (2003) Zones of approach for craniofacial resection: minimizing facial incisions for resection of anterior cranial base and paranasal sinus tumors. Neurosurgery 53(5):1126–1135; discussion 1135–1127. https://doi.org/10.1227/01.neu.0000088802.58956.5a
Aghdasi N, Whipple M, Humphreys IM, Moe KS, Hannaford B, Bly RA (2018) Automated surgical approach planning for complex skull base targets: development and validation of a cost function and semantic at-las. Surg Innov 25(5):476–484. https://doi.org/10.1177/1553350618782287
Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV (2009) Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46(3):786–802. https://doi.org/10.1016/j.neuroimage.2008.12.037
Iosifescu DV, Shenton ME, Warfield SK, Kikinis R, Dengler J, Jolesz FA, McCarley RW (1997) An automated registration algorithm for measuring MRI subcortical brain structures. Neuroimage 6(1):13–25. https://doi.org/10.1006/nimg.1997.0274
Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A (2006) Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage 33(1):115–126. https://doi.org/10.1016/j.neuroimage.2006.05.061
Fatyga M, Dogan N, Weiss E, Sleeman WCT, Zhang B, Lehman WJ, Williamson JF, Wijesooriya K, Christensen GE (2015) A voxel-by-voxel comparison of deformable vector fields obtained by three deformable image registration algorithms applied to 4DCT lung studies. Front Oncol 5:17. https://doi.org/10.3389/fonc.2015.00017
Peroni M, Spadea MF, Riboldi M, Falcone S, Vaccaro C, Sharp GC, Baroni G (2013) Validation of automatic contour propagation for 4D treatment planning using multiple metrics. Technol Cancer Res Treat 12(6):501–510. https://doi.org/10.7785/tcrt.2012.500347
Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ (1999) Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging 18(8):712–721. https://doi.org/10.1109/42.796284
Keeve E, Girod S, Kikinis R, Girod B (1998) Deformable modeling of facial tissue for craniofacial surgery simulation. Comput Aided Surg 3(5):228–238. https://doi.org/10.1002/(sici)1097-0150(1998)3:5%3c228::Aid-igs2%3e3.0.Co;2-i
Klein S, Staring M, Murphy K, Viergever MA, Pluim JP (2010) elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29(1):196–205. https://doi.org/10.1109/TMI.2009.2035616
Pierson RJH, Harris G, Keefe H, Paulsen J, Andreasen N, Magnotta V (2011) Fully automated analysis using BRAINS: AutoWorkUp. Neuroimage 54:328–336
Zaffino P, Raudaschl P, Fritscher K, Sharp GC, Spadea MF (2016) Technical note: plastimatch mabs, an open source tool for automatic image segmentation. Med Phys 43(9):5155. https://doi.org/10.1118/1.4961121
Teguh DN, Levendag PC, Voet PW, Al-Mamgani A, Han X, Wolf TK, Hibbard LS, Nowak P, Akhiat H, Dirkx ML, Heijmen BJ, Hoogeman MS (2011) Clinical validation of atlas-based auto-segmentation of multiple target volumes and normal tissue (swallowing/mastication) structures in the head and neck. Int J Radiat Oncol Biol Phys 81(4):950–957. https://doi.org/10.1016/j.ijrobp.2010.07.009
Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D (2009) Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage 46(3):726–738. https://doi.org/10.1016/j.neuroimage.2009.02.018
Raudaschl PF, Zaffino P, Sharp GC, Spadea MF, Chen A, Dawant BM, Albrecht T, Gass T, Langguth C, Luthi M, Jung F, Knapp O, Wesarg S, Mannion-Haworth R, Bowes M, Ashman A, Guillard G, Brett A, Vincent G, Orbes-Arteaga M, Cardenas-Pena D, Castellanos-Dominguez G, Aghdasi N, Li Y, Berens A, Moe K, Hannaford B, Schubert R, Fritscher KD (2017) Evaluation of segmentation methods on head and neck CT: Auto-segmentation challenge 2015. Med Phys 44(5):2020–2036. https://doi.org/10.1002/mp.12197
Avants BB, Tustison N, Song G (2009) Advanced normalization tools (ANTS). Insight j 2:1–35
Murphy K, van Ginneken B, Reinhardt JM, Kabus S, Ding K, Deng X, Cao K, Du K, Christensen GE, Garcia V, Vercauteren T, Ayache N, Commowick O, Malandain G, Glocker B, Paragios N, Navab N, Gorbunova V, Sporring J, de Bruijne M, Han X, Heinrich MP, Schnabel JA, Jenkinson M, Lorenz C, Modat M, McClelland JR, Ourselin S, Muenzing SE, Viergever MA, De Nigris D, Collins DL, Arbel T, Peroni M, Li R, Sharp GC, Schmidt-Richberg A, Ehrhardt J, Werner R, Smeets D, Loeckx D, Song G, Tustison N, Avants B, Gee JC, Staring M, Klein S, Stoel BC, Urschler M, Werlberger M, Vandemeulebroucke J, Rit S, Sarrut D, Pluim JP (2011) Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. IEEE Trans Med Imaging 30(11):1901–1920. https://doi.org/10.1109/TMI.2011.2158349
Fedorov ABR, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward SR, Miller JV, Pieper S, Kikinis R (2012) 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30(9):1323–1341
Taha AA, Hanbury A (2015) Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 15:29. https://doi.org/10.1186/s12880-015-0068-x
Zou KHWS, Bharatha A, Tempany CM, Kaus MR, Haker SJ, Wells WM 3rd, Jolesz FA, Kikinis R (2004) Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol 11(2):178–189
Bartko JJ (1991) Measurement and reliability: statistical thinking considerations. Schizophr Bull 17(3):483–489. https://doi.org/10.1093/schbul/17.3.483
Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC (1994) Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans Med Imaging 13(4):716–724. https://doi.org/10.1109/42.363096
Sharp G, Fritscher KD, Pekar V, Peroni M, Shusharina N, Veeraraghavan H, Yang J (2014) Vision 20/20: perspectives on automated image segmentation for radiotherapy. Med Phys 41(5):050902. https://doi.org/10.1118/1.4871620
Powell KA, Liang T, Hittle B, Stredney D, Kerwin T, Wiet GJ (2017) Atlas-based segmentation of temporal bone anatomy. Int J Comput Assist Radiol Surg 12(11):1937–1944. https://doi.org/10.1007/s11548-017-1658-6
Damopoulos D, Lerch TD, Schmaranzer F, Tannast M, Chênes C, Zheng G, Schmid J. Segmentation of the proximal femur in radial MR scans using a random forest classifier and deformable model registration.
Zhang Z, Zhao T, Gay H, Zhang W, Sun B (2021) ARPM-net: a novel CNN-based adversarial method with Markov random field enhancement for prostate and organs at risk segmentation in pelvic CT images. Med Phys 48(1):227–237. https://doi.org/10.1002/mp.14580
Pinter C, Lasso A, Wang A, Jaffray D, Fichtinger G (2012) SlicerRT: radiation therapy research toolkit for 3D Slicer. Med Phys 39(10):6332–6338. https://doi.org/10.1118/1.4754659
Schreier J, Genghi A, Laaksonen H, Morgas T, Haas B (2020) Clinical evaluation of a full-image deep segmentation algorithm for the male pelvis on cone-beam CT and CT. Radiother Oncol 145:1–6. https://doi.org/10.1016/j.radonc.2019.11.021
Nowinski WL (2017) 3D Atlas of the brain, head and neck in 2953 pieces. Neuroinformatics 15(4):395–400. https://doi.org/10.1007/s12021-017-9339-8
Schiemann T, Freudenberg J, Pflesser B, Pommert A, Priesmeyer K, Riemer M, Schubert R, Tiede U, Hohne KH (2000) Exploring the visible human using the VOXEL-MAN framework. Comput Med Imaging Graph 24(3):127–132
Iacono MI, Neufeld E, Akinnagbe E, Bower K, Wolf J, Vogiatzis Oikonomidis I, Sharma D, Lloyd B, Wilm BJ, Wyss M, Pruessmann KP, Jakab A, Makris N, Cohen ED, Kuster N, Kainz W, Angelone LM (2015) MIDA: a multimodal imaging-based detailed anatomical model of the human head and neck. PLoS ONE 10(4):e0124126. https://doi.org/10.1371/journal.pone.0124126
Commowick O, Gregoire V, Malandain G (2008) Atlas-based delineation of lymph node levels in head and neck computed tomography images. Radiother Oncol 87(2):281–289. https://doi.org/10.1016/j.radonc.2008.01.018
Sjoberg C, Lundmark M, Granberg C, Johansson S, Ahnesjo A, Montelius A (2013) Clinical evaluation of multi-atlas based segmentation of lymph node regions in head and neck and prostate cancer patients. Radiat Oncol 8:229. https://doi.org/10.1186/1748-717X-8-229
Shamonin DP, Bron EE, Lelieveldt BP, Smits M, Klein S, Staring M, Alzheimer’s Disease Neuroimaging I (2013) Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Front Neuroinform 7:50. https://doi.org/10.3389/fninf.2013.00050
Citardi MJ, Batra PS (2007) Intraoperative surgical navigation for endoscopic sinus surgery: rationale and indications. Curr Opin Otolaryngol Head Neck Surg 15(1):23–27. https://doi.org/10.1097/MOO.0b013e3280123130
Schipaanboord B, Boukerroui D, Peressutti D, van Soest J, Lustberg T, Kadir T, Dekker A, van Elmpt W, Gooding M (2019) Can atlas-based auto-segmentation ever be perfect? Insights from extreme value theory. IEEE Trans Med Imaging 38(1):99–106. https://doi.org/10.1109/TMI.2018.2856464
Manlove AE, Romeo G, Venugopalan SR (2020) Craniofacial growth: current theories and influence on management. Oral Maxillofac Surg Clin North Am 32(2):167–175. https://doi.org/10.1016/j.coms.2020.01.007
Nazib A, Fookes C, Perrin D (2018) A comparative analysis of registration tools: traditional vs deep learning approach on high resolution tissue cleared data. arXiv:abs/1810.08315
Tappeiner E, Proll S, Honig M, Raudaschl PF, Zaffino P, Spadea MF, Sharp GC, Schubert R, Fritscher K (2019) Multi-organ segmentation of the head and neck area: an efficient hierarchical neural networks approach. Int J Comput Assist Radiol Surg 14(5):745–754. https://doi.org/10.1007/s11548-019-01922-4
Zaffino P, Ciardo D, Raudaschl P, Fritscher K, Ricotti R, Alterio D, Marvaso G, Fodor C, Baroni G, Amato F, Orecchia R, Jereczek-Fossa BA, Sharp GC, Spadea MF (2018) Multi atlas based segmentation: should we prefer the best atlas group over the group of best atlases? Phys Med Biol 63(12):12NT01. https://doi.org/10.1088/1361-6560/aac712
Moccia S, Foti S, Routray A, Prudente F, Perin A, Sekula RF, Mattos LS, Balzer JR, Fellows-Mayle W, De Momi E, Riviere CN (2018) Toward improving safety in neurosurgery with an active handheld instrument. Ann Biomed Eng 46(10):1450–1464. https://doi.org/10.1007/s10439-018-2091-x
Acknowledgements
NK was supported by T32 DC000018-34 from the National Institute on Deafness and Other Communication Disorders awarded to the University of Washington Department of Otolaryngology (P.I., Edward Weaver). RB was supported by Clinical Research Scholars Program, Center for Clinical and Translational Research, Seattle Children’s Hospital. The authors are grateful to Dr. Ian Humphreys for his help with the clinical validation scoring and Kathryn Whitlock, MS for the statistical power analysis.
Funding
This study was funded by the T32 grant DC000018-34 from the National Institute on Deafness and Other Communication Disorders and the Clinical Research Scholars Program grant from Center for Clinical and Translational Research at Seattle Children’s Hospital.
Author information
Authors and Affiliations
Contributions
NK conceived and designed the analysis, contributed data or analysis tools, performed data analysis, and wrote the paper; FP contributed data or analysis tools and edited the paper; AMM contributed data or analysis tools and edited the paper; WMA assisted in validation and edited the paper; KM assisted in validation and edited the paper; BH conceived and designed the analysis, contributed data or analysis tools, and edited the paper; RB conceived and designed the analysis, contributed data or analysis tools, assisted in validation, and edited the paper.
Corresponding author
Ethics declarations
Conflict of Interest
NK declares that they have no conflict of interest. FAP declares that they have no conflict of interest. AMM declares that they have no conflict of interest. WMA declares that they have no conflict of interest. KM is a cofounder of SpiWay, LLC. BH declares that they have no conflict of interest. RB is a cofounder of EigenHealth, Inc., a consultant to SpiWay, LLC, and holds a financial interest of ownership equity with Edus Health, Inc.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
The study was approved by Seattle Children’s Hospital Institutional Review Board (SCH IRB # STUDY00001830).
Informed Consent
Retrospective study: For this type of study formal consent is not required.
Code availability
Data available for download http://hdl.handle.net/1773/46259.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Konuthula, N., Perez, F.A., Maga, A.M. et al. Automated atlas-based segmentation for skull base surgical planning. Int J CARS 16, 933–941 (2021). https://doi.org/10.1007/s11548-021-02390-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02390-5