Abstract
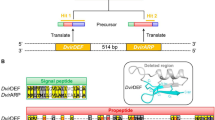
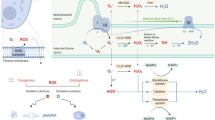
Autophagy is an evolutionarily conserved process in eukaryotes, which is regulated by autophagy-related genes (ATGs). Arthrobotrys oligospora is a representative species of nematode-trapping (NT) fungi that can produce special traps for nematode predation. To elucidate the biological roles of autophagy in NT fungi, we characterized an orthologous Atg protein, AoAtg5, in A. oligospora. We found that AoATG5 deletion causes a significant reduction in vegetative growth and conidiation, and that the transcript levels of several sporulation-related genes were significantly downregulated during sporulation stage. In addition, the cell nuclei were significantly reduced in the ΔAoATG5 mutant, and the transcripts of several genes involved in DNA biosynthesis, repair, and ligation were significantly upregulated. In ΔAoATG5 mutants, the autophagic process was significantly impaired, and trap formation and nematocidal activity were significantly decreased. Comparative transcriptome analysis results showed that AoAtg5 is involved in the regulation of multiple cellular processes, such as autophagy, nitrogen metabolism, DNA biosynthesis and repair, and vesicular transport. In summary, our results suggest that AoAtg5 is essential for autophagy and significantly contributes to vegetative growth, cell nucleus development, sporulation, trap formation, and pathogenicity in A. oligospora, thus providing a basis for future studies focusing on related mechanisms of autophagy in NT fungi.
Similar content being viewed by others
References
Chen, Y.L., Gao, Y., Zhang, K.Q., and Zou, C.G. (2013). Autophagy is required for trap formation in the nematode-trapping fungus Arthrobotrys oligospora. Environ Microbiol Rep 5, 511–517.
Dijksterhuis, J., Veenhuis, M., Harder, W., and Nordbring-Hertz, B. (1994). Nematophagous fungi: physiological aspects and structure-function relationships. Adv Microb Physiol 36, 111–143.
Farré, J.C., and Subramani, S. (2016). Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol 17, 537–552.
Fujioka, Y., Noda, N.N., Fujii, K., Yoshimoto, K., Ohsumi, Y., and Inagaki, F. (2008). In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J Biol Chem 283, 1921–1928.
Füllgrabe, J., Lynch-Day, M.A., Heldring, N., Li, W., Struijk, R.B., Ma, Q., Hermanson, O., Rosenfeld, M.G., Klionsky, D.J., and Joseph, B. (2013). The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500, 468–471.
Füllgrabe, J., Klionsky, D.J., and Joseph, B. (2014). The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 15, 65–74.
Gatica, D., Lahiri, V., and Klionsky, D.J. (2018). Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20, 233–242.
Guzder, S.N., Sung, P., Bailly, V., Prakash, L., and Prakash, S. (1994). RAD25 is a DMA helicase required for DNA repair and RNA polymerase II transcription. Nature 369, 578–581.
Hanada, T., Noda, N.N., Satomi, Y., Ichimura, Y., Fujioka, Y., Takao, T., Inagaki, F., and Ohsumi, Y. (2007). The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282, 37298–37302.
Itakura, E., and Mizushima, N. (2010). Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776.
Ji, X., Li, H., Zhang, W., Wang, J., Liang, L., Zou, C., Yu, Z., Liu, S., and Zhang, K.Q. (2020). The lifestyle transition of Arthrobotrys oligospora is mediated by microRNA-like RNAs. Sci China Life Sci 63, 543–551.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870–1874.
Lai, Y., Chen, H., Wei, G., Wang, G., Li, F., and Wang, S. (2017). In vivo gene expression profiling of the entomopathogenic fungus Beauveria bassiana elucidates its infection stratagems in Anopheles mosquito. Sci China Life Sci 60, 839–851.
Li, B., and Dewey, C.N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf 12, 323.
Liu, N., Ning, G.A., Liu, X.H., Feng, X.X., Lu, J.P., Mao, L.J., Su, Z.Z., Wang, Y., Zhang, C.L., and Lin, F.C. (2016). An autophagy gene, HoATG5, is involved in sporulation, cell wall integrity and infection of wounded barley leaves. Microbiol Res 192, 326–335.
Liu, X.H., Yang, J., He, R.L., Lu, J.P., Zhang, C.L., Lu, S.L., and Lin, F.C. (2011). An autophagy gene, TrATG5, affects conidiospore differentiation in Trichoderma reesei. Res Microbiol 162, 756–763.
Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408.
Lu, J.P., Liu, X.H., Feng, X.X., Min, H., and Lin, F.C. (2009). An autophagy gene, MgATG5, is required for cell differentiation and pathogenesis in Magnaporthe oryzae. Curr Genet 55, 461–473.
Lv, W., Wang, C., Yang, N., Que, Y., Talbot, N.J., and Wang, Z. (2017). Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci Rep 7, 11062.
Ma, Y., Yang, X., Xie, M., Zhang, G., Yang, L., Bai, N., Zhao, Y., Li, D., Zhang, K.Q., and Yang, J. (2020). The Arf-GAP AoGlo3 regulates conidiation, endocytosis, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Fungal Genets Biol 138, 103352.
Maresca, T.J., Freedman, B.S., and Heald, R. (2005). Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol 169, 859–869.
Maskey, D., Yousefi, S., Schmid, I., Zlobec, I., Perren, A., Friis, R., and Simon, H.U. (2013). ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun 4, 2130.
Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178.
Nordbring-Hertz, B. (2004). Morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora—an extensive plasticity of infection structures. Mycologist 18, 125–133.
Park, H.S., Bayram, O., Braus, G.H., Kim, S.C., and Yu, J.H. (2012). Characterization of the velvet regulators in Aspergillus fumigatus. Mol Microbiol 86, 937–953.
Pollack, J.K., Harris, S.D., and Marten, M.R. (2009). Autophagy in filamentous fungi. Fungal Genets Biol 46, 1–8.
Shi, T.Q., Liu, G.N., Ji, R.Y., Shi, K., Song, P., Ren, L.J., Huang, H., and Ji, X.J. (2017). CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl Microbiol Biotechnol 101, 7435–7443.
Simon, H.U., and Friis, R. (2014). ATG5: A distinct role in the nucleus. Autophagy 10, 176–177.
Song, Q., and Kumar, A. (2012). An overview of autophagy and yeast pseudohyphal growth: integration of signaling pathways during nitrogen stress. Cells 1, 263–283.
Su, H., Zhao, Y., Zhou, J., Feng, H., Jiang, D., Zhang, K.Q., and Yang, J. (2017). Trapping devices of nematode-trapping fungi: formation, evolution, and genomic perspectives. Biol Rev 92, 357–368.
Suzuki, K., and Ohsumi, Y. (2007). Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett 581, 2156–2161.
Talbot, N.J., Ebbole, D.J., and Hamer, J.E. (1993). Identification and characterization of Mpg1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5, 1575.
Trapnell, C., Pachter, L., and Salzberg, S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111.
Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333, 169–174.
Tunlid, A., Ãhman, J., and Oliver, R.P. (1999). Transformation of the nematode-trapping fungus Arthrobotrys oligospora. FEMS Microbiol Lett 173, 111–116.
Wang, Y.Y., Zhang, X.Y., Zhou, Q.M., Zhang, X.L., and Wei, J.C. (2015). Comparative transcriptome analysis of the lichen-forming fungus Endocarpon pusillum elucidates its drought adaptation mechanisms. Sci China Life Sci 58, 89–100.
Xie, M., Wang, Y., Tang, L., Yang, L., Zhou, D., Li, Q., Niu, X., Zhang, K. Q., and Yang, J. (2019). AoStuA, an APSES transcription factor, regulates the conidiation, trap formation, stress resistance and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. Environ Microbiol 21, 4648–4661.
Xie, M., Bai, N., Yang, J., Jiang, K., Zhou, D., Zhao, Y., Li, D., Niu, X., Zhang, K.Q., and Yang, J. (2020). Protein kinase Ime2 is required for mycelial growth, conidiation, osmoregulation, and pathogenicity in nematode-trapping fungus Arthrobotrys oligospora. Front Microbiol 10, 3065.
Yang, C.T., Vidal-Diez de Ulzurrun, G., Gonçalves, A.P., Lin, H.C., Chang, C.W., Huang, T.Y., Chen, S.A., Lai, C.K., Tsai, I.J., Schroeder, F.C., et al. (2020). Natural diversity in the predatory behavior facilitates the establishment of a robust model strain for nematode-trapping fungi. Proc Natl Acad Sci USA 117, 6762–6770.
Yang, J., Wang, L., Ji, X., Feng, Y., Li, X., Zou, C., Xu, J., Ren, Y., Mi, Q., Wu, J., et al. (2011). Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog 7, e1002179.
Yang, J., Yu, Y., Li, J., Zhu, W., Geng, Z., Jiang, D., Wang, Y., and Zhang, K.Q. (2013). Characterization and functional analyses of the chitinase-encoding genes in the nematode-trapping fungus Arthrobotrys oligospora. Arch Microbiol 195, 453–462.
Yang, X., Ma, N., Yang, L., Zheng, Y., Zhen, Z., Li, Q., Xie, M., Li, J., Zhang, K.Q., and Yang, J. (2018). Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl Microbiol Biotechnol 102, 4601–4613.
Ying, S.H., and Feng, M.G. (2019). Insight into vital role of autophagy in sustaining biological control potential of fungal pathogens against pest insects and nematodes. Virulence 10, 429–437.
Zhang, G., Zheng, Y., Ma, Y., Yang, L., Xie, M., Zhou, D., Niu, X., Zhang, K.Q., and Yang, J. (2019). The velvet proteins VosA and VelB play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front Microbiol 10, 1917.
Zhang, L., Wang, J., Xie, X.Q., Keyhani, N.O., Feng, M.G., and Ying, S.H. (2013). The autophagy gene BbATG5, involved in the formation of the autophagosome, contributes to cell differentiation and growth but is dispensable for pathogenesis in the entomopathogenic fungus Beauveria bassiana. Microbiology 159, 243–252.
Zhen, Z., Xing, X., Xie, M., Yang, L., Yang, X., Zheng, Y., Chen, Y., Ma, N., Li, Q., Zhang, K.Q., et al. (2018). MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genets Biol 116, 42–50.
Zhen, Z., Zhang, G., Yang, L., Ma, N., Li, Q., Ma, Y., Niu, X., Zhang, K.Q., and Yang, J. (2019). Characterization and functional analysis of calcium/calmodulin-dependent protein kinases (CaMKs) in the nematode-trapping fungus Arthrobotrys oligospora. Appl Microbiol Biotechnol 103, 819–832.
Zhou, D., Xie, M., Bai, N., Yang, L., Zhang, K.Q., and Yang, J. (2020). The autophagy-related gene Aolatg4 regulates hyphal growth, sporulation, autophagosome formation, and pathogenicity in Arthrobotrys oligospora. Front Microbiol 11, 59224.
Zhu, X.M., Li, L., Wu, M., Liang, S., Shi, H.B., Liu, X.H., and Lin, F.C. (2018). Current opinions on autophagy in pathogenicity of fungi. Virulence 10, 481–489.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31960556 and U1402265), the Applied Basic Research Foundation of Yunnan Province (202001BB050004), and the National Basic Research Program of China (2013CB127503). We would be grateful to Guo Yingqi (Kunming Institute of Zoology, Chinese Academy of Sciences) for her help of taking and analyzing EM images.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Compliance and ethics The author(s) declare that they have no conflict of interest.
Supporting Information
Rights and permissions
About this article
Cite this article
Zhou, D., Zhu, Y., Bai, N. et al. AoATG5 plays pleiotropic roles in vegetative growth, cell nucleus development, conidiation, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 65, 412–425 (2022). https://doi.org/10.1007/s11427-020-1913-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-020-1913-9