Abstract
Purpose
To compare values of multiparametric magnetic resonance imaging (MRI) sequences and propose clinical-radiomics nomogram for diagnosis of soft-tissue sarcoma (STS).
Procedures
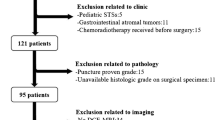
This study enrolled 148 patients from Dec. 2017 to Feb. 2021. All patients underwent T1-weighted (T1W), contrast-enhanced T1-weighted (T1CE), and T2-weighted fat-suppressed (T2FS) MRI scans. A total of 1967 radiomic features were extracted from the segmented regions of interest (ROIs) in each MRI sequence. Highly diagnostic radiomic features were selected with Mann–Whitney U test, elastic net, and Akaike’s information criterion (AIC) based on MRI images. Logistical regression was used to build Rad scores. Clinical factors were analyzed using the chi-square test or Mann–Whitney U test. The performance of the Rad scores was judged using the area under the receiver operating characteristic area under the curve (ROC AUC), sensitivity, specificity, and accuracy. The nomogram was developed by integrating the Rad score and the most important clinical factor.
Results
By combining the three MRI sequences, the Rad-Com was developed consisting of twelve features selected by with Mann–Whitney U test, elastic net, and AIC: four from T1W, three from TICE, and five from T2FS MRI. The margin (P < 0.05) demonstrated a statistically significant difference between patients with benign and malignant soft-tissue tumors (STT). The nomogram was constructed by integrating the Rad-Com and margin, which yielded favorable diagnostic AUCs of 0.919 (sensitivity (Sen) = 0.784, specificity (Spe) = 0.936) and 0.913 (Sen = 0.923, Spe = 0.792) in the training and validation cohort.
Conclusion
The proposed nomogram may have potential as a noninvasive marker for STS diagnosis.







Similar content being viewed by others
Change history
30 August 2022
A Correction to this paper has been published: https://doi.org/10.1007/s11307-022-01761-x
References
Honoré C, Faron M, Mir O et al (2018) Management of locoregional recurrence after radical resection of a primary nonmetastatic retroperitoneal soft tissue sarcoma: the Gustave Roussy experience. J Surg Oncol 118:1318–1325. https://doi.org/10.1002/jso.25291
Gamboa AC, Gronchi A, Cardona K (2020) Soft-tissue sarcoma in adults: an update on the current state of histiotype-specific management in an era of personalized medicine. CA A Cancer J Clin 70:200–229. https://doi.org/10.3322/caac.21605
Issels RD, Lindner LH, Verweij J et al (2018) Effect of neoadjuvant chemotherapy plus regional hyperthermia on long-term outcomes among patients with localized high-risk soft tissue sarcoma: the EORTC 62961-ESHO 95 Randomized Clinical Trial. JAMA Oncol 4:483. https://doi.org/10.1001/jamaoncol.2017.4996
Demetri GD, Baker LH, Beech D et al (2005) Soft tissue sarcoma clinical practice guidelines in oncology. J Natl Compr Canc Netw 3:158–194
Ezuddin NS, Pretell-Mazzini J, Yechieli RL et al (2018) Local recurrence of soft-tissue sarcoma: issues in imaging surveillance strategy. Skeletal Radiol 47:1595–1606. https://doi.org/10.1007/s00256-018-2965-x
Hoang NT, Acevedo LA, Mann MJ, Tolani B (2018) A review of soft-tissue sarcomas: translation of biological advances into treatment measures. CMAR 10:1089–1114. https://doi.org/10.2147/CMAR.S159641
Frassica FJ, Khanna JA, McCarthy EF (2000) The role of MR imaging in soft tissue tumor evaluation. Magn Reson Imaging Clin N Am 8:915–927. https://doi.org/10.1016/S1064-9689(21)00650-4
Chung WJ, Chung HW, Shin MJ et al (2012) MRI to differentiate benign from malignant soft-tissue tumours of the extremities: a simplified systematic imaging approach using depth, size and heterogeneity of signal intensity. BJR 85:e831–e836. https://doi.org/10.1259/bjr/27487871
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762. https://doi.org/10.1038/nrclinonc.2017.141
Kumar V, Gu Y, Basu S et al (2012) Radiomics: the process and the challenges. Magn Reson Imaging 30:1234–1248. https://doi.org/10.1016/j.mri.2012.06.010
Sollini M, Antunovic L, Chiti A, Kirienko M (2019) Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. Eur J Nucl Med Mol Imaging 46:2656–2672. https://doi.org/10.1007/s00259-019-04372-x
Wang H, Chen H, Duan S et al (2020) Radiomics and machine learning with multiparametric preoperative MRI may accurately predict the histopathological grades of soft tissue sarcomas. J Magn Reson Imaging 51:791–797. https://doi.org/10.1002/jmri.26901
Zhang Y, Zhu Y, Shi X et al (2019) Soft tissue sarcomas: preoperative predictive histopathological grading based on radiomics of MRI. Acad Radiol 26:1262–1268. https://doi.org/10.1016/j.acra.2018.09.025
Farhidzadeh H, Goldgof DB, Hall LO, et al (2015) Texture feature analysis to predict metastatic and necrotic soft tissue sarcomas. In: 2015 IEEE International Conference on Systems, Man, and Cybernetics. IEEE, Kowloon Tong, Hong Kong, pp 2798–2802
Kakino R, Nakamura M, Mitsuyoshi T et al (2020) Comparison of radiomic features in diagnostic CT images with and without contrast enhancement in the delayed phase for NSCLC patients. Physica Med 69:176–182. https://doi.org/10.1016/j.ejmp.2019.12.019
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Zou H, Hastie T (2005) Regularization and variable selection via the elastic net. J Royal Statistical Soc B 67:301–320. https://doi.org/10.1111/j.1467-9868.2005.00503.x
Shen C, Liu Z, Wang Z et al (2018) Building CT radiomics based nomogram for preoperative esophageal cancer patients lymph node metastasis prediction. Translational Oncology 11:815–824. https://doi.org/10.1016/j.tranon.2018.04.005
Bozdogan H (1987) Model selection and Akaike’s Information Criterion (AIC): the general theory and its analytical extensions. Psychometrika 52:345–370. https://doi.org/10.1007/BF02294361
Sun R-J, Fang M-J, Tang L et al (2020) CT-based deep learning radiomics analysis for evaluation of serosa invasion in advanced gastric cancer. Eur J Radiol 132:109277. https://doi.org/10.1016/j.ejrad.2020.109277
Wu L, Deng F, Xie Z et al (2016) Spatial analysis of severe fever with thrombocytopenia syndrome virus in China using a geographically weighted logistic regression model. IJERPH 13:1125. https://doi.org/10.3390/ijerph13111125
Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF (2008) Youden index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom J 50:419–430. https://doi.org/10.1002/bimj.200710415
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837. https://doi.org/10.2307/2531595
Fields BKK, Demirjian NL, Hwang DH et al (2021) Whole-tumor 3D volumetric MRI-based radiomics approach for distinguishing between benign and malignant soft tissue tumors. Eur Radiol 31:8522–8535. https://doi.org/10.1007/s00330-021-07914-w
Morii T, Kishino T, Shimamori N et al (2018) Differential diagnosis between benign and malignant soft tissue tumors utilizing ultrasound parameters. J Med Ultrasonics 45:113–119. https://doi.org/10.1007/s10396-017-0796-3
Choi YJ, Lee IS, Song YS et al (2019) Diagnostic performance of diffusion-weighted (DWI) and dynamic contrast-enhanced (DCE) MRI for the differentiation of benign from malignant soft-tissue tumors: diagnostic performance of DWI and DCE. J Magn Reson Imaging 50:798–809. https://doi.org/10.1002/jmri.26607
Wang H, Nie P, Wang Y et al (2020) Radiomics nomogram for differentiating between benign and malignant soft-tissue masses of the extremities. J Magn Reson Imaging 51:155–163. https://doi.org/10.1002/jmri.26818
Lee SE, Jung J-Y, Nam Y et al (2021) Radiomics of diffusion-weighted MRI compared to conventional measurement of apparent diffusion-coefficient for differentiation between benign and malignant soft tissue tumors. Sci Rep 11:15276. https://doi.org/10.1038/s41598-021-94826-w
Juntu J, Sijbers J, De Backer S et al (2010) Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J Magn Reson Imaging 31:680–689. https://doi.org/10.1002/jmri.22095
Xu R, Kido S, Suga K et al (2014) Texture analysis on 18F-FDG PET/CT images to differentiate malignant and benign bone and soft-tissue lesions. Ann Nucl Med 28:926–935. https://doi.org/10.1007/s12149-014-0895-9
Malinauskaite I, Hofmeister J, Burgermeister S et al (2020) Radiomics and machine learning differentiate soft-tissue lipoma and liposarcoma better than musculoskeletal radiologists. Sarcoma 2020:1–9. https://doi.org/10.1155/2020/7163453
Peeken JC, Asadpour R, Specht K et al (2021) MRI-based delta-radiomics predicts pathologic complete response in high-grade soft-tissue sarcoma patients treated with neoadjuvant therapy. Radiother Oncol 164:73–82. https://doi.org/10.1016/j.radonc.2021.08.023
Molina D, Pérez-Beteta J, Luque B et al (2016) Tumour heterogeneity in glioblastoma assessed by MRI texture analysis: a potential marker of survival. BJR 89:20160242. https://doi.org/10.1259/bjr.20160242
Chitalia RD, Kontos D (2019) Role of texture analysis in breast MRI as a cancer biomarker: a review. J Magn Reson Imaging 49:927–938. https://doi.org/10.1002/jmri.26556
Li Z, Mao Y, Huang W et al (2017) Texture-based classification of different single liver lesion based on SPAIR T2W MRI images. BMC Med Imaging 17:42. https://doi.org/10.1186/s12880-017-0212-x
Funding
The study was supported by China National Natural Science Foundation (31770147), Project of Pneumoconiosis Prevention and Control of China's coal mines foundation (201909J033) and Natural Science Foundation of Liaoning Province (2021-MS-205).
Author information
Authors and Affiliations
Contributions
Zhibin Yue: conceptualization, methodology, software, writing — original draft, visualization. Xiaoyu Wang: data curation, methodology, writing — review and editing, visualization. Yan Wang: visualization, writing — original draft, validation, investigation. Hongbo Wang: investigation, supervision, software, review and editing. Wenyan Jiang: validation, writing — review and editing, supervision, conceptualization, methodology.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article was updated to add the equal contribution note missed during the production process.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yue, Z., Wang, X., Wang, Y. et al. Clinical-Radiomics Nomogram from T1W, T1CE, and T2FS MRI for Improving Diagnosis of Soft-Tissue Sarcoma. Mol Imaging Biol 24, 995–1006 (2022). https://doi.org/10.1007/s11307-022-01751-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-022-01751-z