Abstract
Purpose
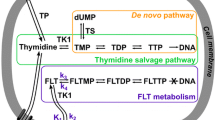
The primary goal of this study is to evaluate the accuracy of the fluorescence ubiquitination cell cycle indicator (FUCCI) system with fluorescence in vivo imaging compared to 3′-deoxy-3′-[18F]fluorothymidine ([18F]-FLT) positron emission tomography (PET)/computed tomography (CT) and biological validation through histology. Imaging with [18F]-FLT PET/CT can be used to noninvasively assess cancer cell proliferation and has been utilized in both preclinical and clinical studies. However, a cost-effective and straightforward method for in vivo, cell cycle targeted cancer drug screening is needed prior to moving towards translational imaging methods such as PET/CT.
Procedures
In this study, fluorescent MDA-MB-231-FUCCI tumor growth was monitored weekly with caliper measurements and fluorescent imaging. Seven weeks post-injection, [18F]-FLT PET/CT was performed with a preclinical PET/CT, and tumors samples were harvested for histological analysis.
Results
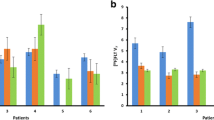
RFP fluorescent signal significantly correlated with tumor volume (r = 0.8153, p < 0.0001). Cell proliferation measured by GFP fluorescent imaging was correlated with tumor growth rate (r = 0.6497, p < 0.001). Also, GFP+ cells and [18F]-FLT regions of high uptake were both spatially located in the tumor borders, indicating that the FUCCI-IVIS method may provide an accurate assessment of tumor heterogeneity of cell proliferation. The quantification of total GFP signal was correlated with the sum of tumor [18F]-FLT standard uptake value (SUV) (r = 0.5361, p = 0.0724). Finally, histological analysis confirmed viable cells in the tumor and the correlation of GFP + and Ki67 + cells (r = 0.6368, p = 0.0477).
Conclusion
Fluorescent imaging of the cell cycle provides a noninvasive accurate depiction of tumor progression and response to therapy, which may benefit in vivo testing of novel cancer therapeutics that target the cell cycle.




Similar content being viewed by others
References
Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M (2013) Targeting cell cycle regulation in cancer therapy. Pharmacol Ther 138(2):255–271
Schwartz GK, Shah MA (2005) Targeting the cell cycle: a new approach to cancer therapy. J Clin Oncol Off J Am Soc Clin Oncol 23(36):9408–9421
Sherr CJ, Bartek J (2017) Cell cycle–targeted cancer therapies. Annual Rev Cancer Biol 1(1):41–57
Samadi P, Saki S, Dermani FK, Pourjafar M, Saidijam M (2018) Emerging ways to treat breast cancer: will promises be met? Cell Oncol (Dordr) 41(6):605–621
McCann KE, Hurvitz SA, McAndrew N (2019) Advances in targeted therapies for triple-negative breast cancer. Drugs 79(11):1217–1230
Nishimura M, Onoe T, Sakai H, Arase M, Watanabe S, Soyama M et al (2019) Safety and relative dose intensity of dose-dense doxorubicin and cyclophosphamide followed by dose-dense paclitaxel. Anticancer Res 39(8):4379–4383
Schneeweiss A, Mobus V, Tesch H, Hanusch C, Denkert C, Lubbe K et al (2019) Intense dose-dense epirubicin, paclitaxel, cyclophosphamide versus weekly paclitaxel, liposomal doxorubicin (plus carboplatin in triple-negative breast cancer) for neoadjuvant treatment of high-risk early breast cancer (GeparOcto-GBG 84): a randomised phase III trial. Eur J Cancer (Oxford, England: 1990) 106:181–92
Eikesdal HP, Yndestad S, Elzawahry A, Llop-Guevara A, Gilje B, Blix ES et al (2021) Olaparib monotherapy as primary treatment in unselected triple negative breast cancer. Ann Oncol 32(2):240–249
Marra A, Trapani D, Viale G, Criscitiello C, Curigliano G (2020) Practical classification of triple-negative breast cancer: intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer 6:54
Schmid P, Abraham J, Chan S, Wheatley D, Brunt AM, Nemsadze G et al (2020) Capivasertib Plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer: the PAKT trial. J Clin Oncol 38(5):423–433
Mikhaeel NG (2006) Use of FDG-PET to monitor response to chemotherapy and radiotherapy in patients with lymphomas. Eur J Nucl Med Mol Imaging 33(Suppl 1):22–26
Elmi A, Makvandi M, Weng CC, Hou C, Clark AS, Mach RH et al (2019) Cell-proliferation imaging for monitoring response to CDK4/6 inhibition combined with endocrine-therapy in breast cancer: comparison of [(18)F]FLT and [(18)F]ISO-1 PET/CT. Clin Cancer Res 25(10):3063–3073
Surov A, Meyer HJ, Wienke A (2019) Associations between PET parameters and expression of Ki-67 in breast cancer. Transl Oncol 12(2):375–380
Ducharme M, Fernandez S, Placzek W, Lapi S (2020) Imaging of HER2 expressing tumors with 68Ga radiolabeled peptides. J Nucl Med 61
Lu Y, Li M, Massicano AVF, Song PN, Mansur A, Heinzman KA, et al. (2021) [(89)Zr]-Pertuzumab PET imaging reveals paclitaxel treatment efficacy is positively correlated with HER2 expression in human breast cancer xenograft mouse models. Molecules 26(6)
Hattori Y, Yamasaki T, Ohashi T, Miyanohana Y, Kusumoto T, Maeda R, et al. (2021) Design, synthesis, and evaluation of (11)C-labeled 3-acetyl-indole derivatives as a novel positron emission tomography imaging agent for diacylglycerol kinase gamma (DGKγ) in brain. J Med Chem
Ma G, Liu C, Lian W, Zhang Y, Yuan H, Zhang Y et al (2021) (18)F-FLT PET/CT imaging for early monitoring response to CDK4/6 inhibitor therapy in triple negative breast cancer. Ann Nucl Med 35(5):600–607
Ueberroth BE, Lawhorn-Crews JM, Heilbrun LK, Smith DW, Akoury J, Ali-Fehmi R et al (2019) The use of 3’-deoxy-3’-(18)F-fluorothymidine (FLT) PET in the assessment of long-term survival in breast cancer patients treated with neoadjuvant chemotherapy. Ann Nucl Med 33(6):383–393
Miladinova D (2019) Molecular imaging in breast cancer. Nucl Med Mol Imaging 53(5):313–319
Kramer GM, Liu Y, de Langen AJ, Jansma EP, Trigonis I, Asselin MC et al (2018) Repeatability of quantitative (18)F-FLT uptake measurements in solid tumors: an individual patient data multi-center meta-analysis. Eur J Nucl Med Mol Imaging 45(6):951–961
Leung K (2004) Molecular Imaging and Contrast Agent Database (MICAD). Bethesda: National Center for Biotechnology Information (US)
Pirovano G, Roberts S, Kossatz S, Reiner T (2020) Optical imaging modalities: principles and applications in preclinical research and clinical settings. J Nucl Med 61(10):1419–1427
Müller J, Wunder A, Licha K (2013) Optical imaging. Recent Results Cancer Res 187:221–246
Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H et al (2008) Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132(3):487–498
Zielke N, Edgar BA (2015) FUCCI sensors: powerful new tools for analysis of cell proliferation. Wiley Interdiscip Rev Dev Biol 4(5):469–487
Yano S, Hoffman RM (2018) Real-time determination of the cell-cycle position of individual cells within live tumors using FUCCI cell-cycle imaging. Cells 7(10)
Harris LA, Frick PL, Garbett SP, Hardeman KN, Paudel BB, Lopez CF et al (2016) An unbiased metric of antiproliferative drug effect in vitro. Nat Methods 13(6):497–500
Bloom MJ, Jarrett AM, Triplett TA, Syed AK, Davis T, Yankeelov TE et al (2020) Anti-HER2 induced myeloid cell alterations correspond with increasing vascular maturation in a murine model of HER2+ breast cancer. BMC Cancer 20(1):359
Juríková M, Danihel Ľ, Polák Š, Varga I (2016) Ki67, PCNA, and MCM proteins: markers of proliferation in the diagnosis of breast cancer. Acta Histochem 118(5):544–552
Li LT, Jiang G, Chen Q, Zheng JN (2015) Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol Med Rep 11(3):1566–1572
Menon SS, Guruvayoorappan C, Sakthivel KM, Rasmi RR (2019) Ki-67 protein as a tumour proliferation marker. Clin Chim Acta 491:39–45
Dagogo-Jack I, Shaw AT (2018) Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 15(2):81–94
Roulot A, Héquet D, Guinebretière JM, Vincent-Salomon A, Lerebours F, Dubot C et al (2016) Tumoral heterogeneity of breast cancer. Ann Biol Clin (Paris) 74(6):653–660
Marusyk A, Polyak K (2010) Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 1805(1):105–117
Sanghera B, Wong WL, Sonoda LI, Beynon G, Makris A, Woolf D et al (2014) FLT PET-CT in evaluation of treatment response. Indian J Nucl Med 29(2):65–73
Peck M, Pollack HA, Friesen A, Muzi M, Shoner SC, Shankland EG et al (2015) Applications of PET imaging with the proliferation marker [18F]-FLT. Q J Nucl Med Mol Imaging 59(1):95–104
Guo H, Chen G, Gao M, Wang R, Liu Y, Yu F (2019) Imaging of endogenous hydrogen peroxide during the process of cell mitosis and mouse brain development with a near-infrared ratiometric fluorescent probe. Anal Chem 91(1):1203–1210
Camorani S, Hill BS, Collina F, Gargiulo S, Napolitano M, Cantile M et al (2018) Targeted imaging and inhibition of triple-negative breast cancer metastases by a PDGFRβ aptamer. Theranostics 8(18):5178–5199
Bailly C, Bodet-Milin C, Bourgeois M, Gouard S, Ansquer C, Barbaud M, et al. (2019) Exploring tumor heterogeneity using PET imaging: the big picture. Cancers (Basel) 11(9)
O’Connor JPB (2017) Cancer heterogeneity and imaging. Semin Cell Dev Biol 64:48–57
Mitamura K, Yamamoto Y, Kudomi N, Maeda Y, Norikane T, Miyake K et al (2017) Intratumoral heterogeneity of (18)F-FLT uptake predicts proliferation and survival in patients with newly diagnosed gliomas. Ann Nucl Med 31(1):46–52
Li D, Patel CB, Xu G, Iagaru A, Zhu Z, Zhang L et al (2020) Visualization of diagnostic and therapeutic targets in glioma with molecular imaging. Front Immunol 11:592389
Nishimukai A, Inoue N, Kira A, Takeda M, Morimoto K, Araki K et al (2017) Tumor size and proliferative marker geminin rather than Ki67 expression levels significantly associated with maximum uptake of 18F-deoxyglucose levels on positron emission tomography for breast cancers. PLoS ONE 12(9):e0184508
Sun X, Kaufman PD (2018) Ki-67: more than a proliferation marker. Chromosoma 127(2):175–186
Rivenson Y, Wang H, Wei Z, de Haan K, Zhang Y, Wu Y et al (2019) Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat Biomed Eng 3(6):466–477
Kakimi K, Matsushita H, Hosoi A, Miyai M, Ohara O (2015) CTLs regulate tumor growth via cytostatic effects rather than cytotoxicity: a few T cells can influence the growth of many times more tumor cells. Oncoimmunology 4(3):e970464
Takahashi K, Tanabe R, Ehata S, Kubota SI, Morishita Y, Ueda HR et al (2021) Visualization of the cancer cell cycle by tissue-clearing technology using the Fucci reporter system. Cancer Sci 112(9):3796–3809
Di Blasi R, Marbiah MM, Siciliano V, Polizzi K, Ceroni F (2021) A call for caution in analysing mammalian co-transfection experiments and implications of resource competition in data misinterpretation. Nat Commun 12(1):2545
Jacobsen L, Calvin S, Lobenhofer E (2009) Transcriptional effects of transfection: the potential for misinterpretation of gene expression data generated from transiently transfected cells. Biotechniques 47(1):617–624
Acknowledgements
We acknowledge grant support from the American Cancer Society (RSG-18-006-01-CCE) and National Cancer Institute (R01CA24058) and the O’Neal Comprehensive Cancer Center at UAB’s preclinical shared imaging facility (NIH P30CA013148).
Author information
Authors and Affiliations
Contributions
Conceptualization, A.G.S., J.M.W., and Y.L.; methodology, A.V.F.M.; image analysis, Y.L.; histological staining, Y.L.; histological analysis, C.A.G., K.A.H., and S.W.P.; writing—original draft preparation, Y.L.; writing—review and editing, Y.L., A.V.F.M., C.A.G., K.A.H., S.W.P., J.M.W, and A.G.S.; funding acquisition, A.G.S. All authors have read and agreed to the published version of the manuscript and accountable for all aspects of the work.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, Y., Massicano, A.V.F., Gallegos, C.A. et al. Evaluating the Accuracy of FUCCI Cell Cycle In Vivo Fluorescent Imaging to Assess Tumor Proliferation in Preclinical Oncology Models. Mol Imaging Biol 24, 898–908 (2022). https://doi.org/10.1007/s11307-022-01739-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-022-01739-9